from pyspark.ml.feature import Normalizer normalizer = Normalizer(inputCol='features', outputCol='normalized_features', p=2.0) normalized_df = normalizer.transform(df) normalized_df.show(truncate=False)
+---+--------------+------------------------------------------------------------+ |id |features |normalized_features | +---+--------------+------------------------------------------------------------+ |0 |[1.0,0.1,-1.0]|[0.7053456158585983,0.07053456158585983,-0.7053456158585983]| |1 |[2.0,1.1,1.0] |[0.8025723539051279,0.4414147946478204,0.40128617695256397] | |2 |[3.0,10.1,3.0]|[0.27384986857909926,0.9219612242163009,0.27384986857909926]| +---+--------------+------------------------------------------------------------+
Normalizer scales each row vector (not each column) so that the vector has unit p-norm.
The parameter p controls which type of norm is used.
✅ What is the p-norm?
For a vector:
v=[x1,x2,...,xn]
The p-norm is:

Then Spark normalizes the vector by:

📌 Meaning of different p values
1️⃣ p = 1 → L1 norm
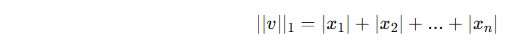
Normalized vector:
