Import Modules:
import numpy as np import pandas as pd import matplotlib.pyplot as plt from sklearn.model_selection import train_test_split from sklearn.model_selection import cross_val_score from sklearn.tree import DecisionTreeClassifier # to build a classification tree from sklearn.tree import plot_tree # to draw a classification tree from sklearn.metrics import confusion_matrix # to create a confusion matrix # from sklearn.metrics import plot_confusion_matrix # deprecated from sklearn.metrics import ConfusionMatrixDisplay # to draw a confusion matrix
Import Data:
# df = pd.read_csv('heart+disease/processed.cleveland.data', header=None) df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/heart-disease/processed.cleveland.data', header=None)
Variables Table
| Variable Name | Role | Type | Demographic | Description | Units | Missing Values |
|---|---|---|---|---|---|---|
| age | Feature | Integer | Age | years | no | |
| sex | Feature | Categorical | Sex | no | ||
| cp | Feature | Categorical | no | |||
| trestbps | Feature | Integer | resting blood pressure (on admission to the hospital) | mm Hg | no | |
| chol | Feature | Integer | serum cholestoral | mg/dl | no | |
| fbs | Feature | Categorical | fasting blood sugar > 120 mg/dl | no | ||
| restecg | Feature | Categorical | no | |||
| thalach | Feature | Integer | maximum heart rate achieved | no | ||
| exang | Feature | Categorical | exercise induced angina | no | ||
| oldpeak | Feature | Integer | ST depression induced by exercise relative to rest | no | ||
| slope | Feature | Categorical | no | |||
| ca | Feature | Integer | number of major vessels (0-3) colored by flourosopy | yes | ||
| thal | Feature | Categorical | yes | |||
| num | Target | Integer | diagnosis of heart disease (the predicted attribute) | no |
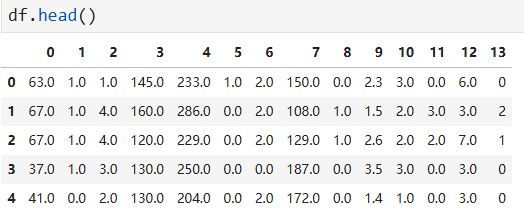
# change the column numbers to column names df.columns = [ 'age', 'sex', 'cp', 'trestbps', 'chol', 'fbs', 'restecg', 'thalach', 'exang', 'oldpeak', 'slope', 'ca', 'thal', 'num' ]
Missing Data Part 1: Identify Missing Data:
df.dtypes

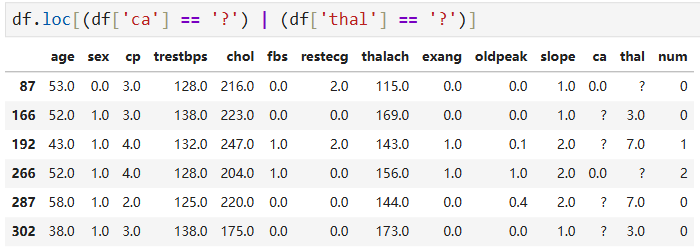
df['ca'].unique() # array(['0.0', '3.0', '2.0', '1.0', '?'], dtype=object) df['thal'].unique() # array(['6.0', '3.0', '7.0', '?'], dtype=object) len(df.loc[(df['ca'] == '?') | (df['thal'] == '?')]) # 6
Missing Data Part 2: Deal with Missing Data:
df_no_missing = df.loc[(df['ca'] != '?') & (df['thal'] != '?')] len(df_no_missing) # 297
Format the Data Part 1: Split the Data into Dependent and Independent Variables
# Make a new copy of the columns used to make prediction X = df_no_missing.drop('num', axis=1).copy() # alternatively: X = df_no_missing.iloc[:, :-1]
# Make a new copy of the column we want to predict y = df_no_missing['num'].copy()

Format the Data Part 2: One-Hot Encoding

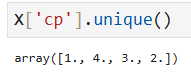

pd.get_dummies(X, columns=['cp']).head()
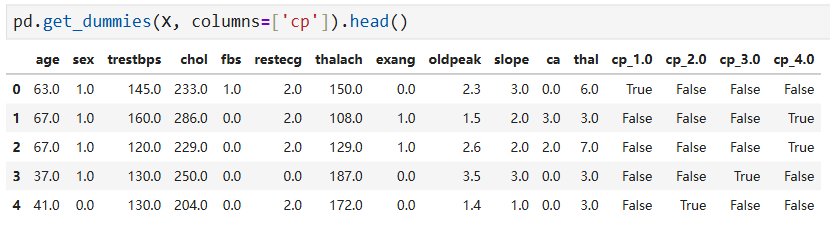
X_encoded = pd.get_dummies(X, columns=['cp', 'restecg', 'slope', 'thal'])
pd.set_option('display.max_columns', None) X_encoded.head()


y_not_zero_index = y > 0 y[y_not_zero_index] = 1 y.unique() # array([0, 1])
Build a Preliminary Classification Tree
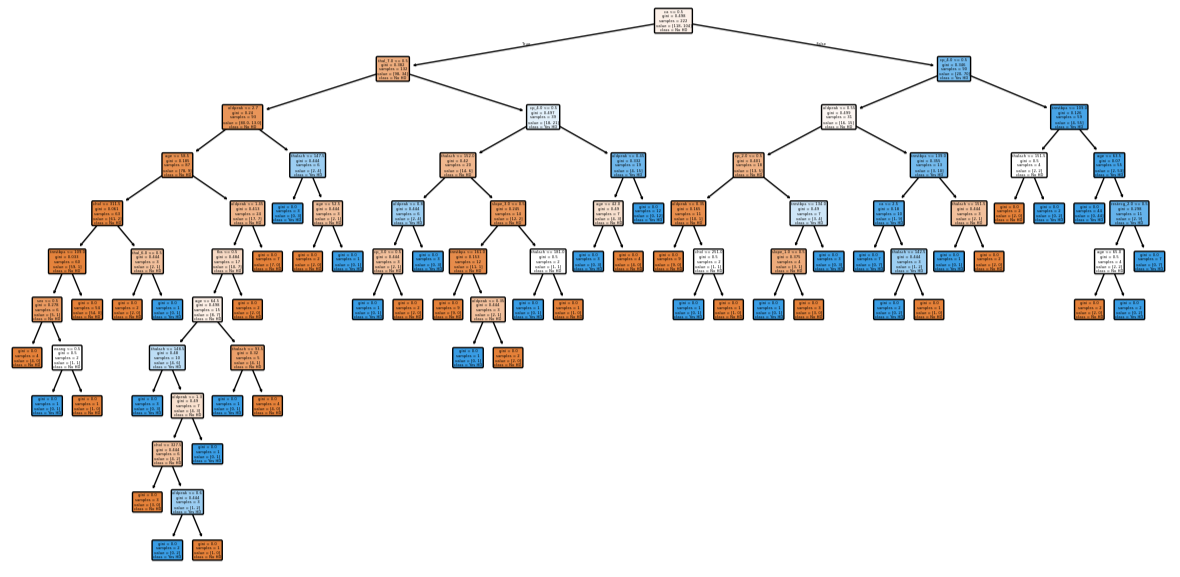
X_train, X_test, y_train, y_test = train_test_split(X_encoded, y, random_state=42) clf_dt = DecisionTreeClassifier(random_state=42) clf_dt = clf_dt.fit(X_train, y_train)
plt.figure(figsize=(15, 7.5)) plot_tree(clf_dt, filled=True, rounded=True, class_names=['No HD', 'Yes HD'], feature_names=X_encoded.columns)
y_pred = clf_dt.predict(X_test) # Compute confusion matrix cm = confusion_matrix(y_test, y_pred) # Display the confusion matrix disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=['Does not have HD', 'Has HD']) disp.plot(cmap='Blues')
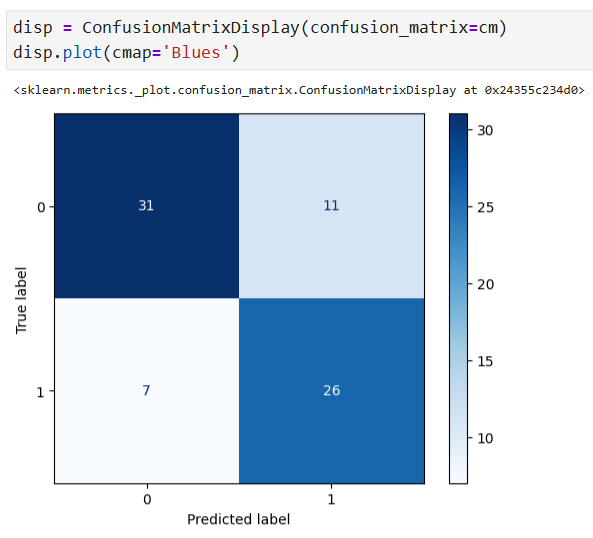
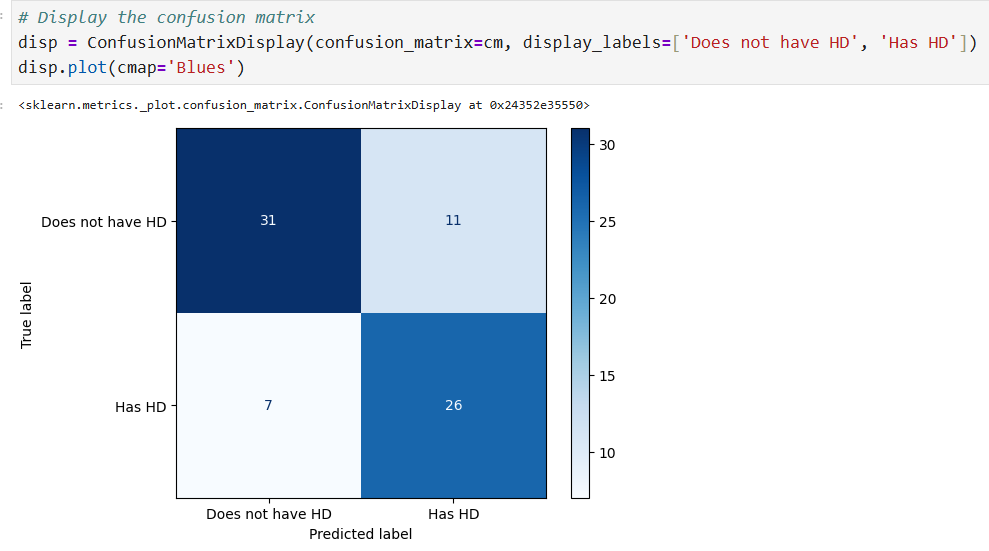
Cost Complexity Pruning Part 1: Visualize alpha

path = clf_dt.cost_complexity_pruning_path(X_train, y_train) # determine values for alpha ccp_alphas = path.ccp_alphas # extract difference values for alpha ccp_alphas = ccp_alphas[:-1] # exclude the maximum value for alpha clf_dts = [] # create an array that we will put decision tress into # create one decision tree per value for alpha and store it in the array for ccp_alpha in ccp_alphas: clf_dt = DecisionTreeClassifier(random_state=42, ccp_alpha=ccp_alpha) clf_dt.fit(X_train, y_train) clf_dts.append(clf_dt)

train_scores = [clf_dt.score(X_train, y_train) for clf_dt in clf_dts] test_scores = [clf_dt.score(X_test, y_test) for clf_dt in clf_dts] fig, ax = plt.subplots() ax.set_xlabel('alpha') ax.set_ylabel('accuracy') ax.set_title('Accuracy vs alpha for training and testing sets') ax.plot(ccp_alphas, train_scores, marker='o', label='train', drawstyle='steps-post') ax.plot(ccp_alphas, test_scores, marker='o', label='test', drawstyle='steps-post') ax.legend() plt.show()
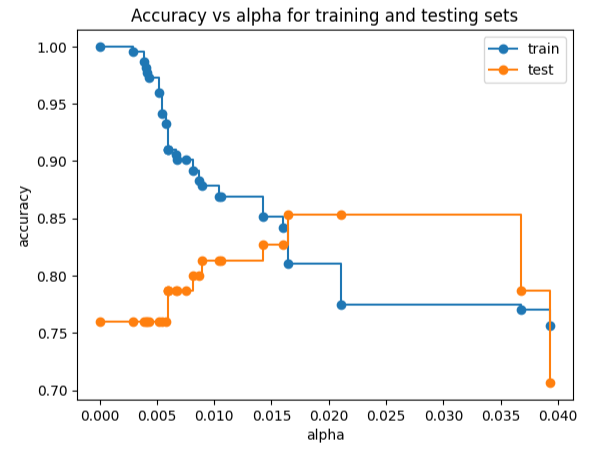

Cost Complexity Pruning Part 2: Cross Validation for Finding the Best Alpha

clf_dt = DecisionTreeClassifier(random_state=42, ccp_alpha=0.016) # create the tree with alpha=0.016 # now use 5-fold cross validation create 5 different training and testing datasets that # are then used to train and test the tree. # NOTE: We use 5-fold because we don't have tons of data. scores = cross_val_score(clf_dt, X_train, y_train, cv=5) df = pd.DataFrame(data={'tree': range(5), 'accuracy': scores}) df.plot(x='tree', y='accuracy', marker='o', linestyle='--')
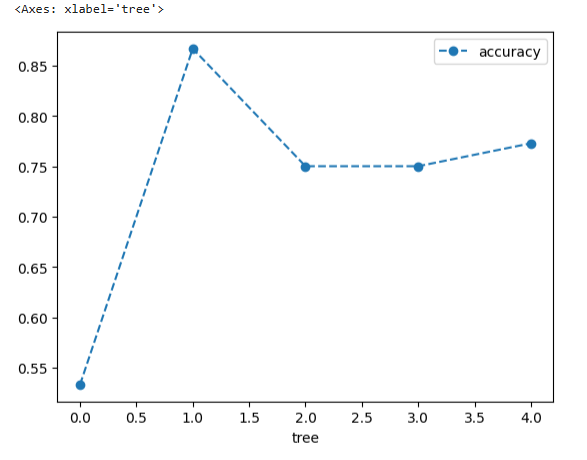

# create an array to store the results of each fold during cross validation alpha_loop_values = [] # For each candidate value for alpha, we will ran 5-fold cross validation # Then we will store the mean and standard deviation of the scores (the accuracy) for each call # to cross_val_score in alpha_loop_values. for ccp_alpha in ccp_alphas: clf_dt = DecisionTreeClassifier(random_state=42, ccp_alpha=ccp_alpha) scores = cross_val_score(clf_dt, X_train, y_train, cv=5) alpha_loop_values.append([ccp_alpha, np.mean(scores), np.std(scores)])
# Now we can draw a graph of the means and standard deviations of the scores # for each candidate value for alpha alpha_results = pd.DataFrame(alpha_loop_values, columns=['alpha', 'accuracy_mean', 'accuracy_std']) alpha_results.plot(x='alpha', y='accuracy_mean', yerr='accuracy_std', marker='o', linestyle='--')
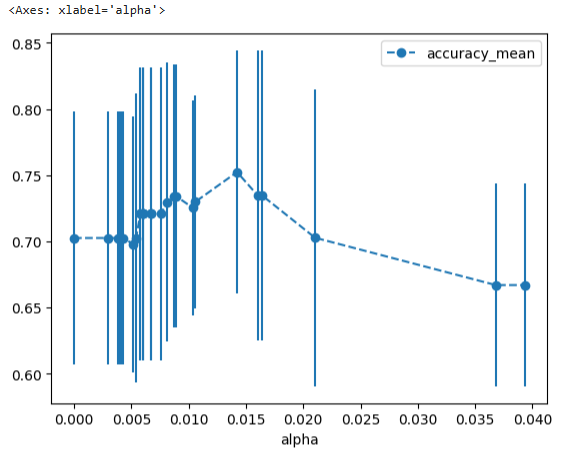

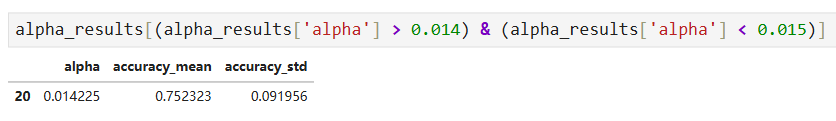
# Store the ideal value for alpha so that we can use it to build the best tree. ideal_ccp_alpha = alpha_results[(alpha_results['alpha'] > 0.014) & (alpha_results['alpha'] < 0.015)]['alpha']
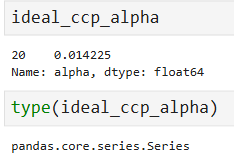

ideal_ccp_alpha = float(ideal_ccp_alpha.iloc[0]) ideal_ccp_alpha # 0.014224751066856332
Build, Evaluate, Draw and Interpret the Final Classification Tree
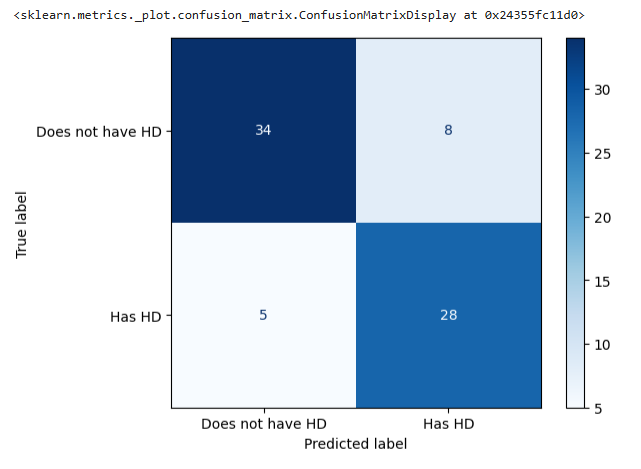
# Draw another confusion matrix to see if the pruned tree does better. y_pred = clf_dt_pruned.predict(X_test) cm = confusion_matrix(y_test, y_pred) disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=['Does not have HD', 'Has HD']) disp.plot(cmap='Blues')

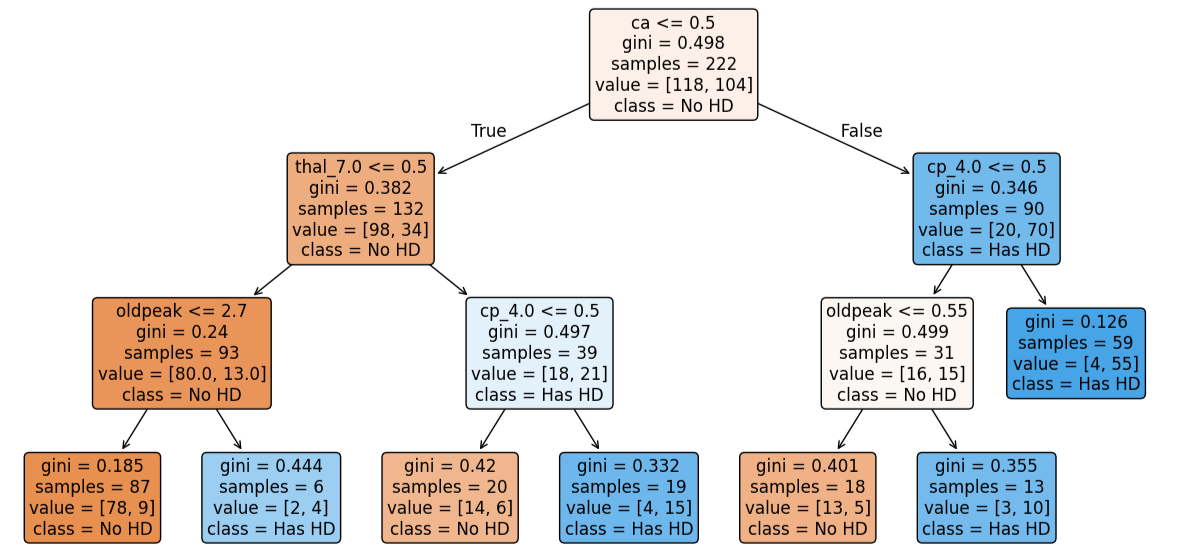
plt.figure(figsize=(15, 7.5)) plot_tree(clf_dt_pruned, filled=True, rounded=True, class_names=['No HD', 'Has HD'], feature_names=X_encoded.columns)




 浙公网安备 33010602011771号
浙公网安备 33010602011771号