

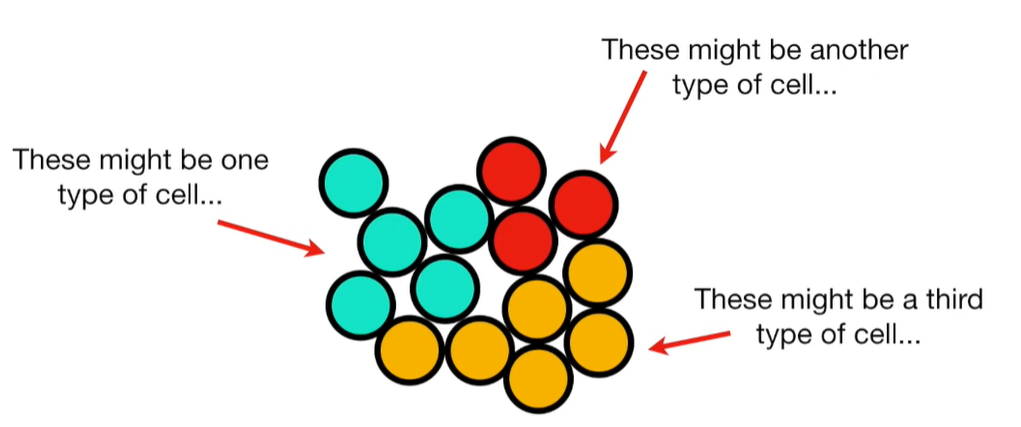

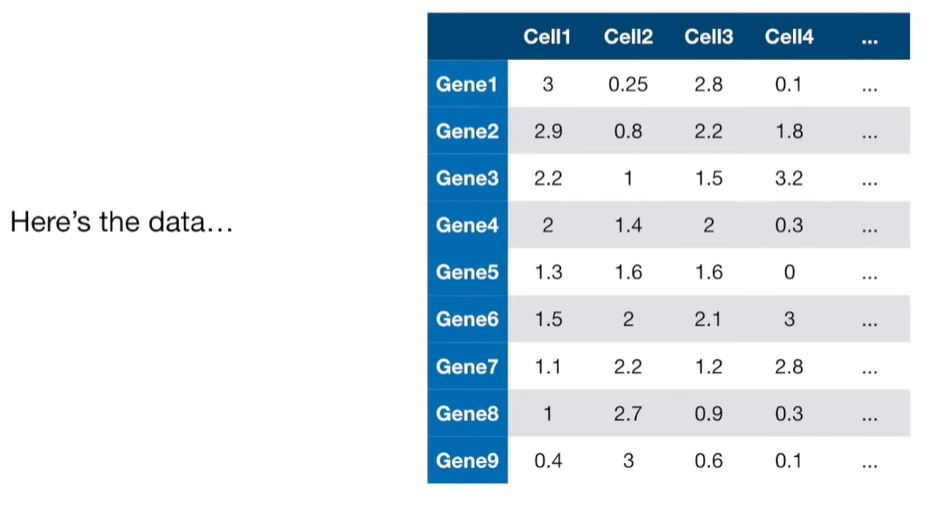
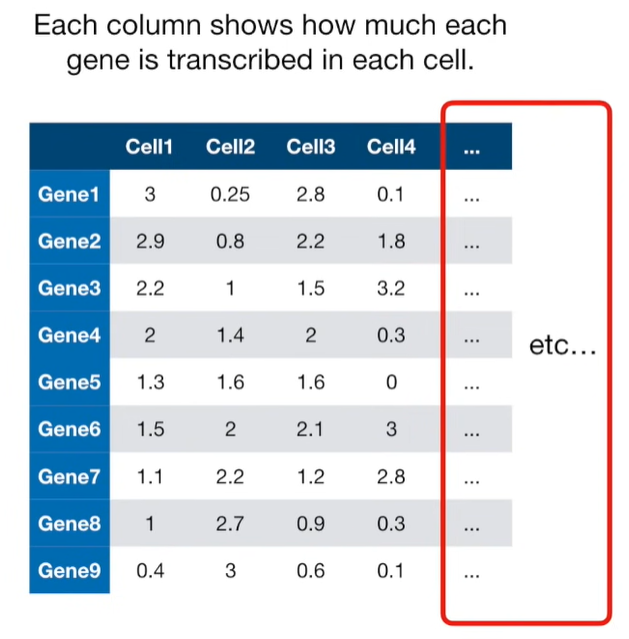
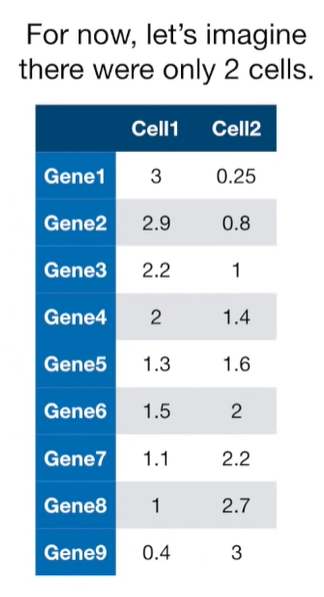
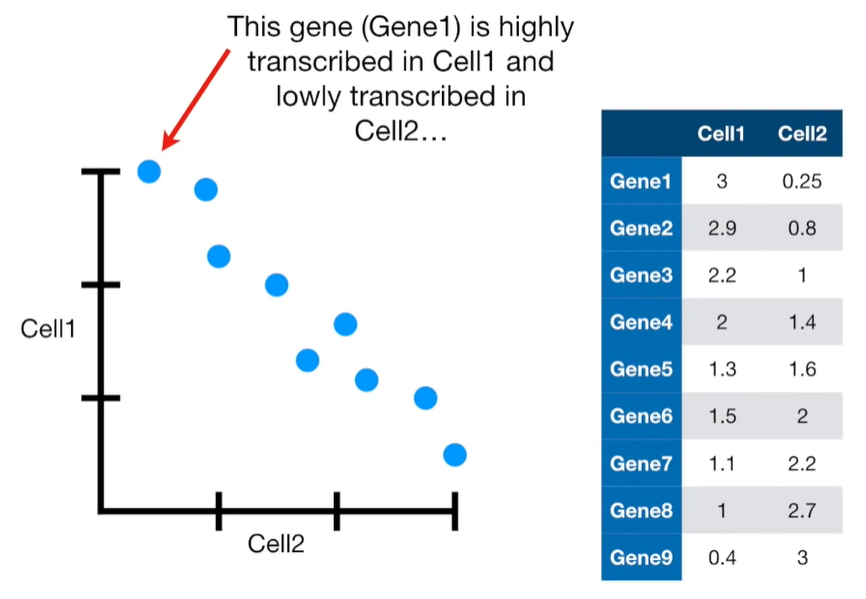
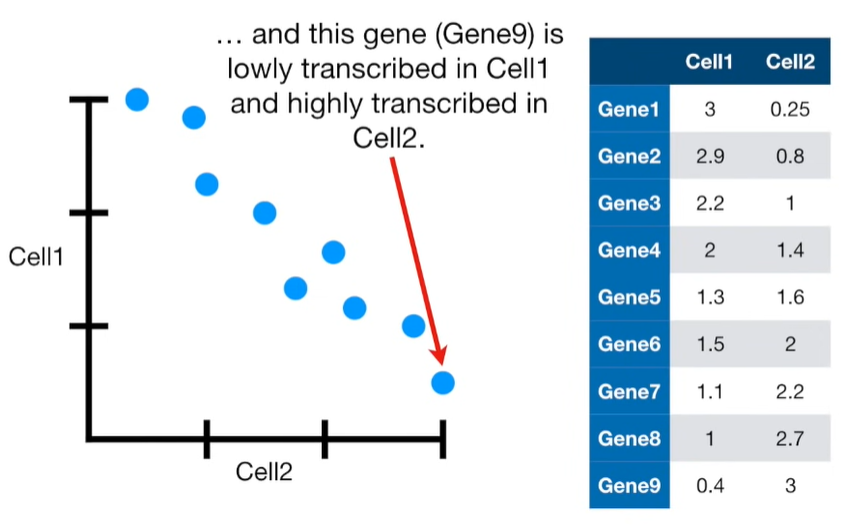
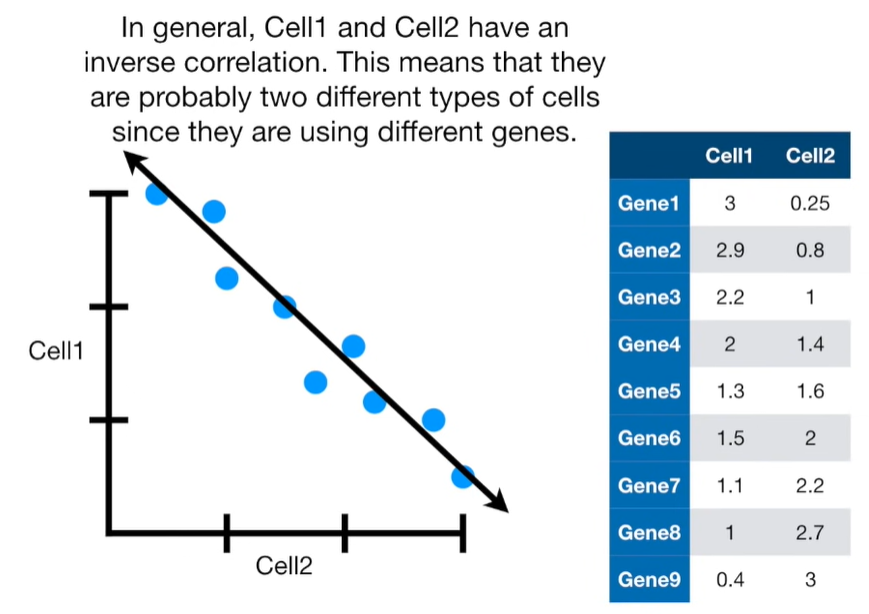
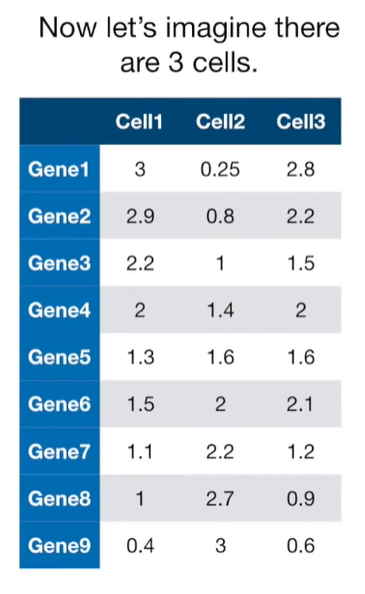

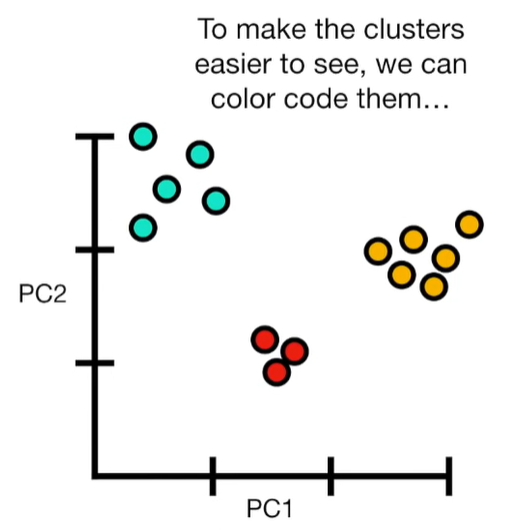
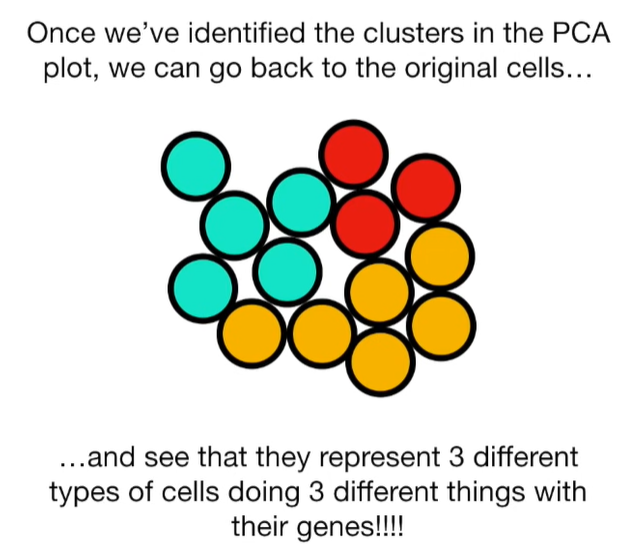
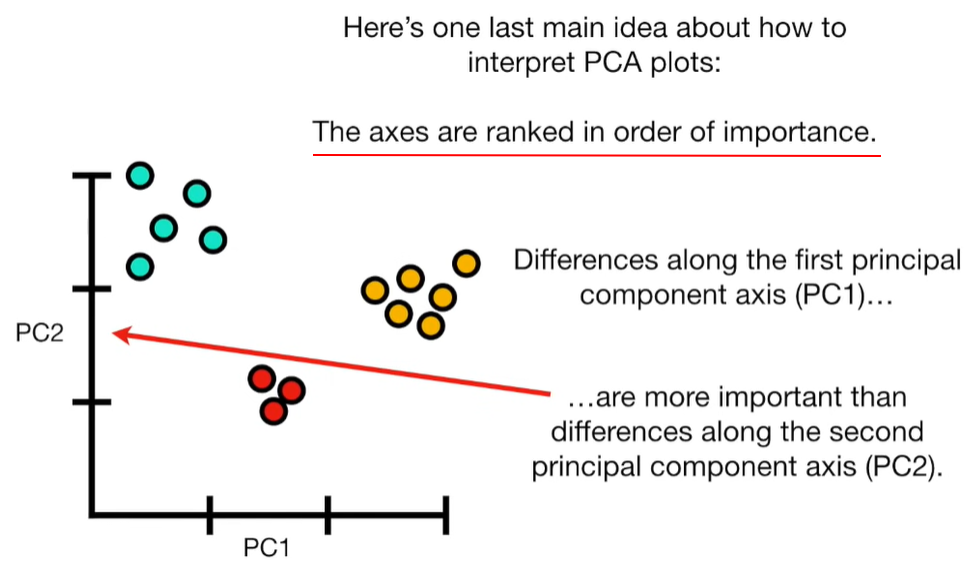
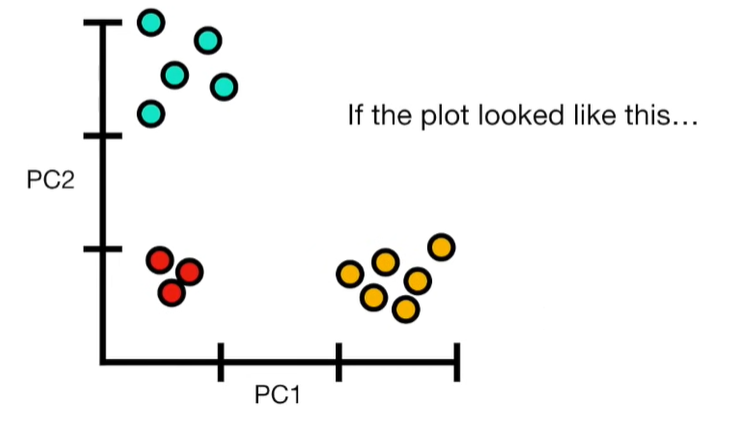




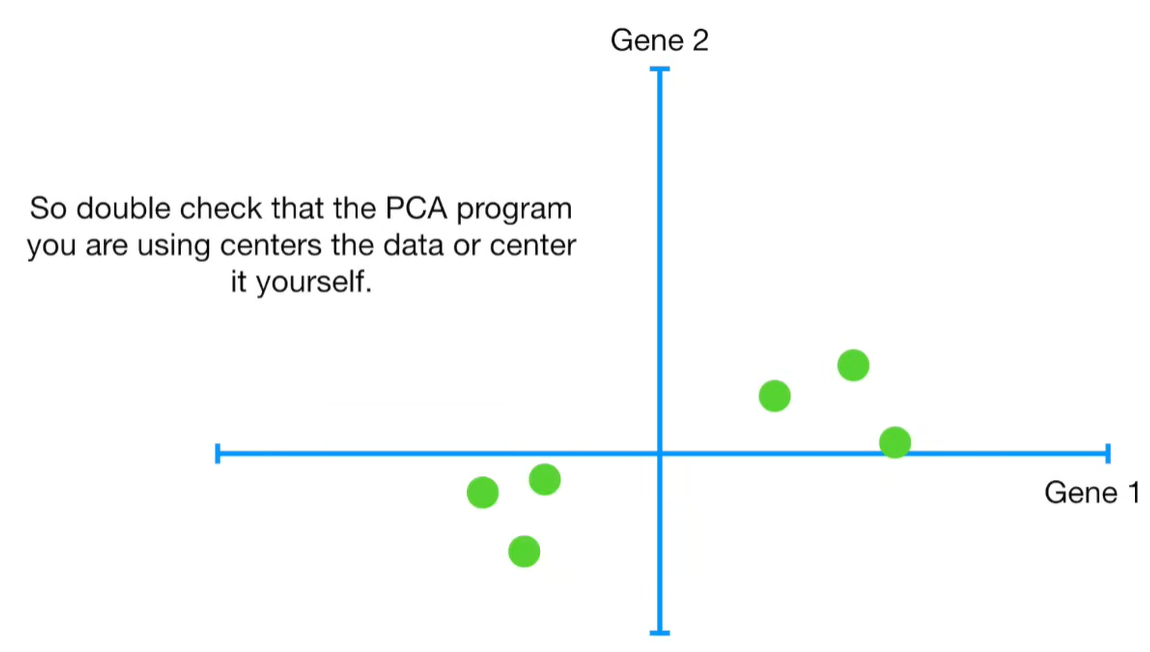

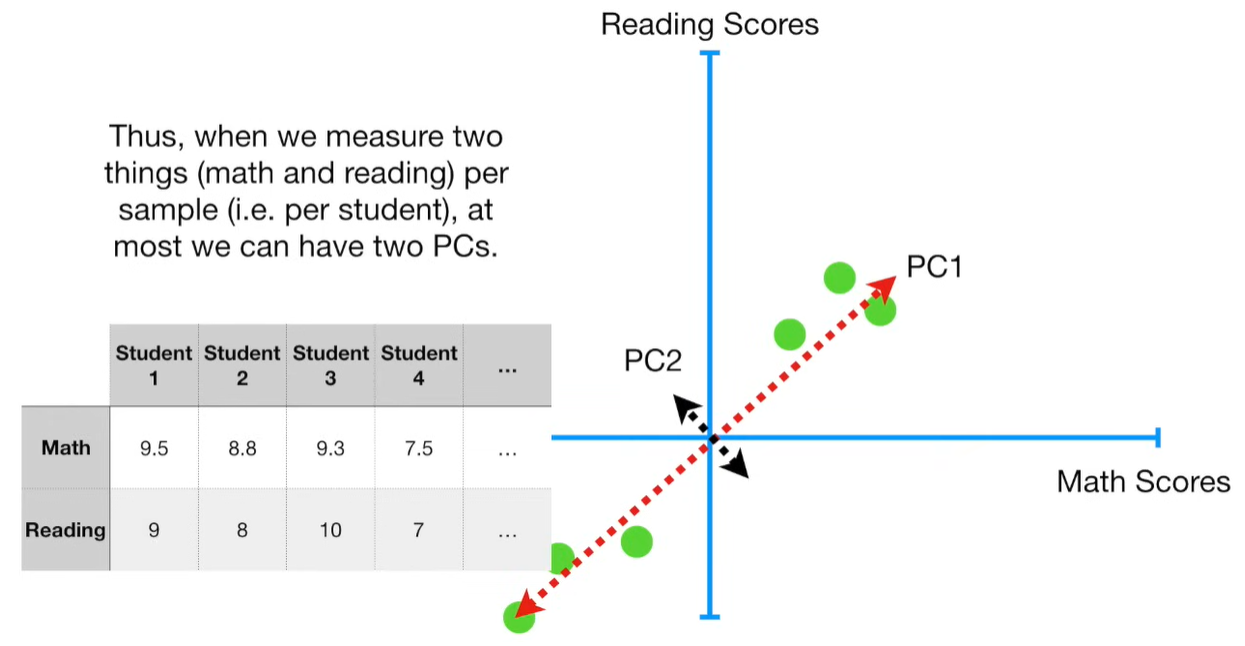
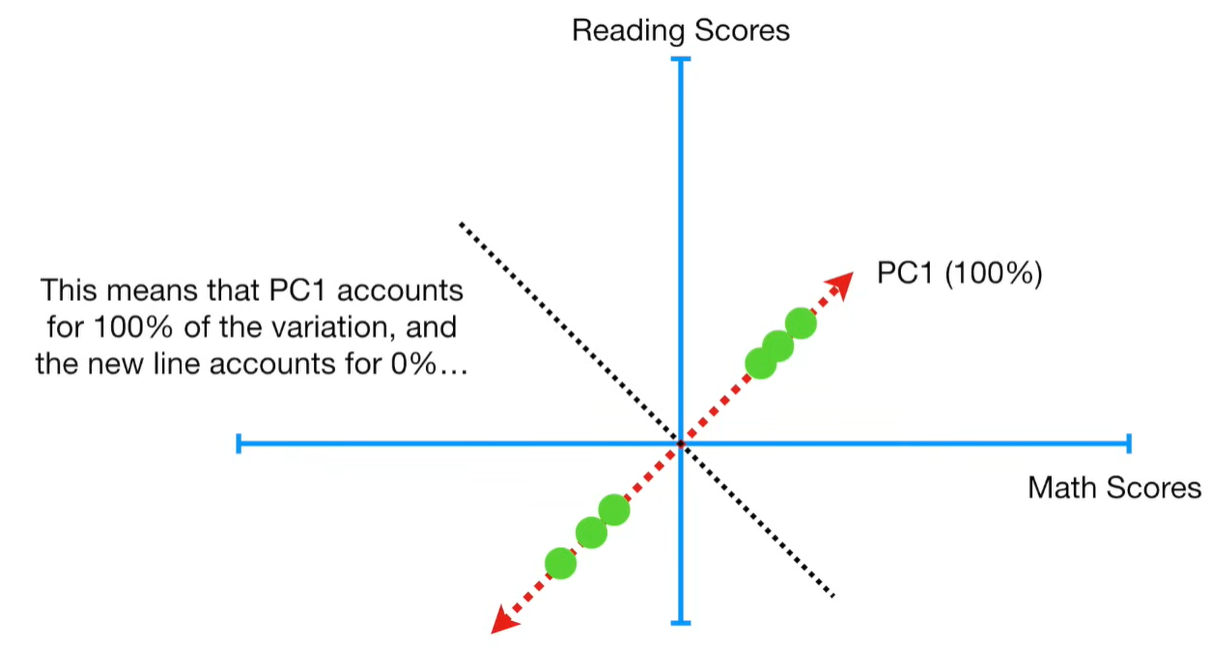
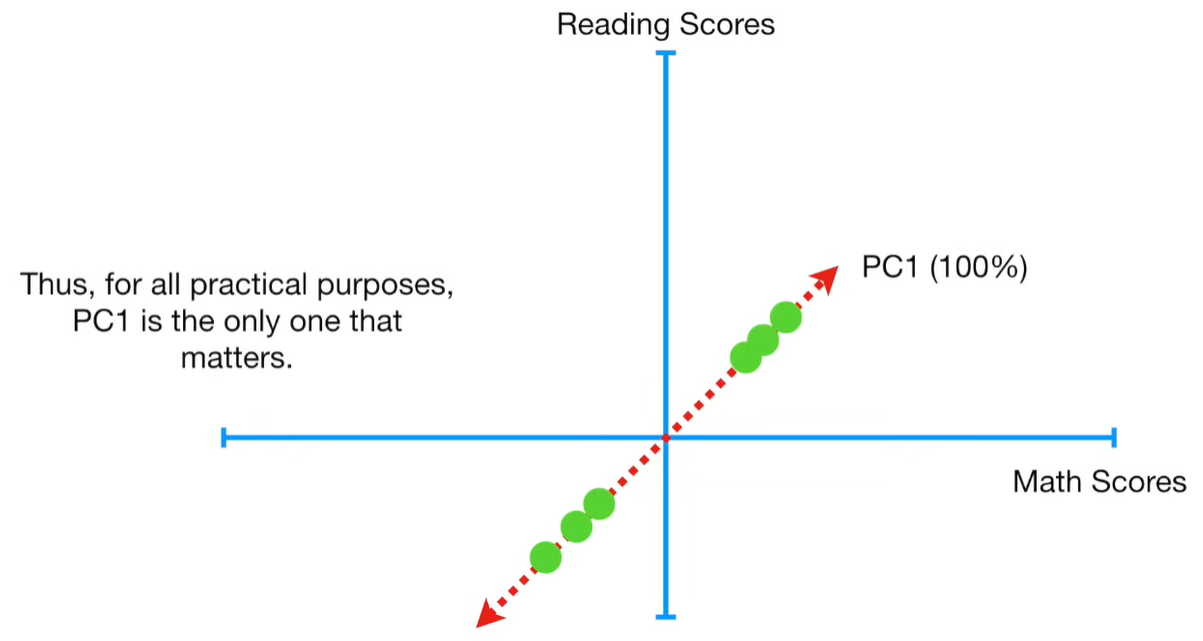



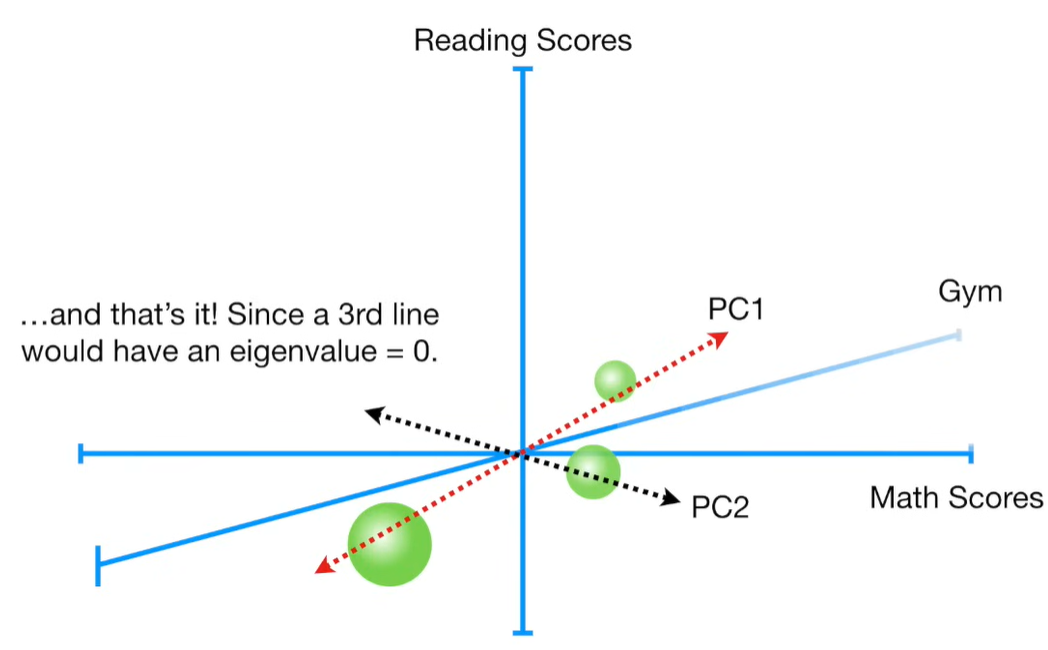
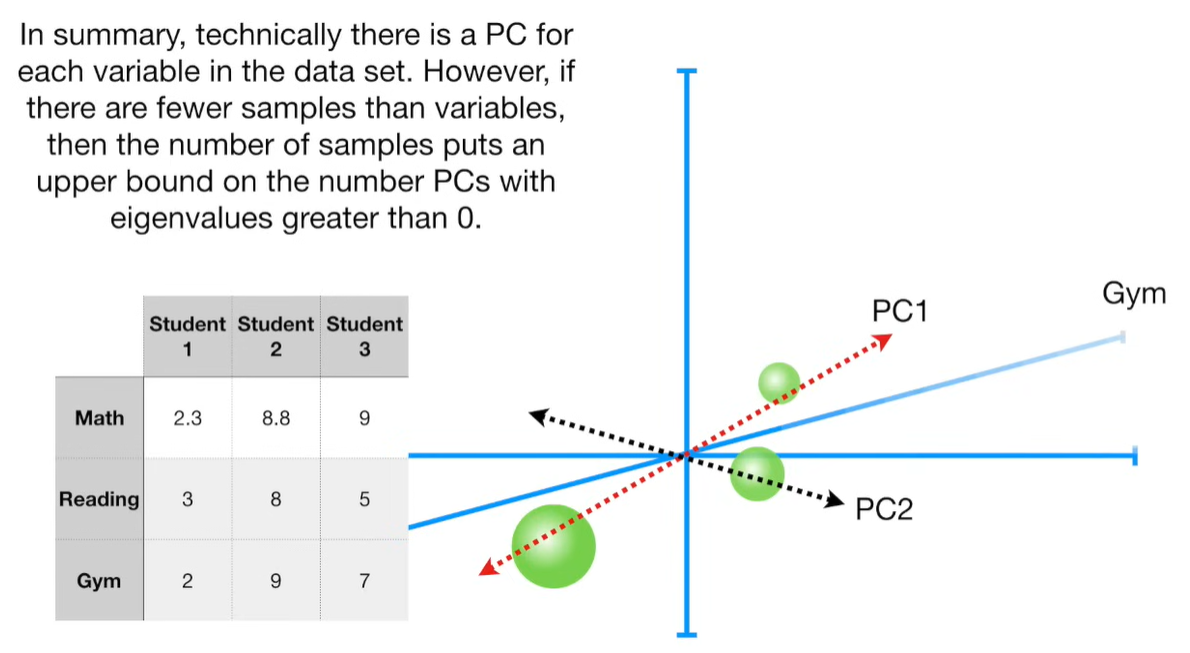
import matplotlib.pyplot as plt import numpy as np import pandas as pd import random as rd from sklearn import preprocessing from sklearn.decomposition import PCA
# features genes = ['gene' + str(i) for i in range (1, 101)] # samples wt = ['wt' + str(i) for i in range(1, 6)] # wild type ko = ['ko' + str(i) for i in range(1, 6)] # knock out data = pd.DataFrame(columns=[*wt, *ko], index=genes) for gene in data.index: data.loc[gene, 'wt1':'wt5'] = np.random.poisson(lam=rd.randrange(10, 1000), size=5) data.loc[gene, 'ko1':'ko5'] = np.random.poisson(lam=rd.randrange(10, 1000), size=5)
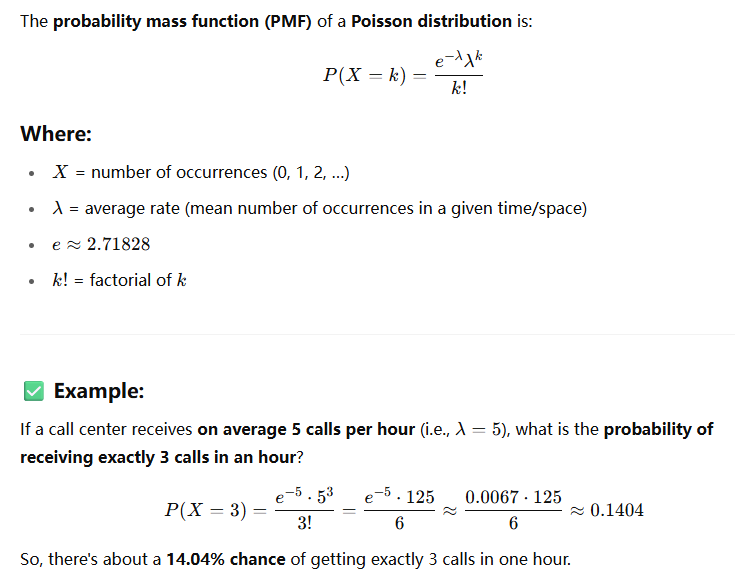 Poisson distribution
Poisson distribution
Here are some classic examples where the Poisson distribution is used to model the number of events happening in a fixed interval of time or space, assuming the events are rare, independent, and occur at a constant average rate:
📦 1. Call Center (Time-Based)
-
Scenario: A call center receives an average of 10 calls per hour.
-
Question: What's the probability of receiving 7 calls in the next hour?
-
Model:
Let X∼Poisson(λ=10)
🚗 2. Traffic Accidents (Time or Space-Based)
-
Scenario: On average, 3 car accidents happen per day on a specific highway.
-
Question: What's the probability of 0 accidents today?
-
Model:
X∼Poisson(λ=3)
📨 3. Emails Received
-
Scenario: You receive about 5 spam emails per day.
-
Question: What’s the probability of getting exactly 8 spam emails tomorrow?
-
Model:
X∼Poisson(λ=5)
🦠 4. Mutations in DNA (Space-Based)
-
Scenario: A particular stretch of DNA has an average of 2 mutations per 1,000 base pairs.
-
Question: What’s the probability of seeing 4 mutations in a 1,000 base pair segment?
-
Model:
X∼Poisson(λ=2)
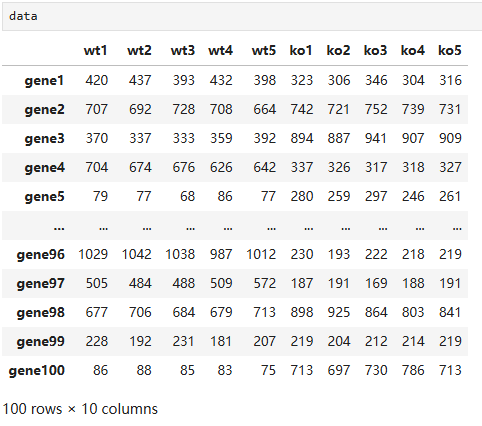
# center and scale the data scaled_data = preprocessing.scale(data.T)



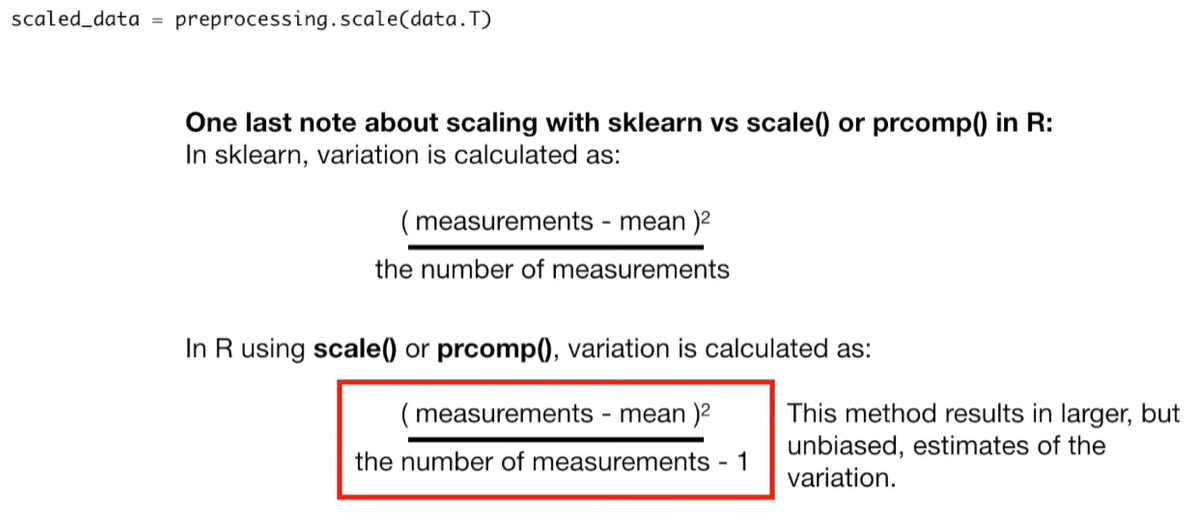


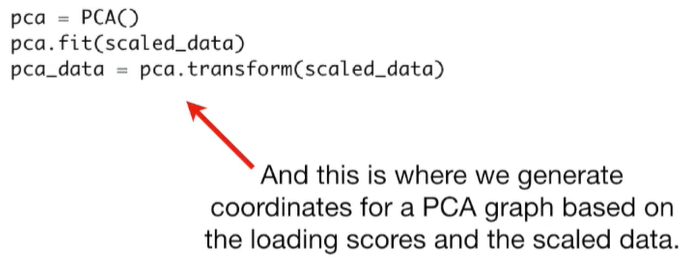
pca = PCA()
pca.fit(scaled_data)
pca_data = pca.transform(scaled_data)

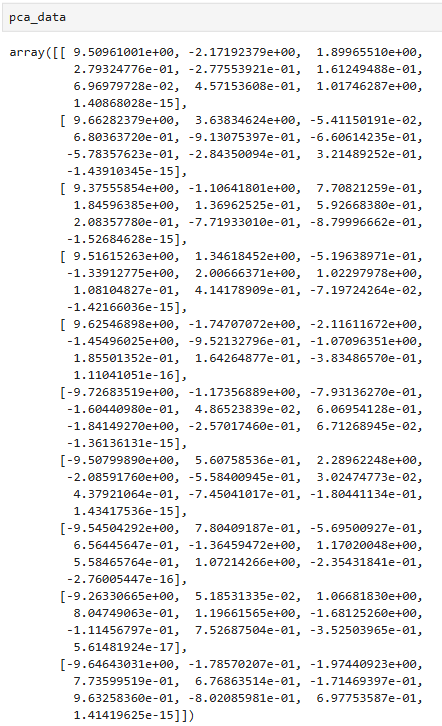
pca_data.shape # (10, 10)
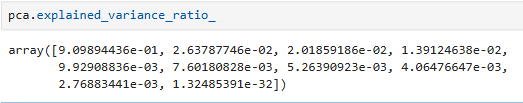
per_var = np.round(pca.explained_variance_ratio_ * 100, decimals=1)

labels = ['PC' + str(i) for i in range(1, len(per_var) + 1)]

plt.bar(x=range(1, len(per_var) + 1), height=per_var, tick_label=labels) plt.title('Scree Plot') plt.xlabel('Principal Component') plt.ylabel('Percentage of Explained Variance') plt.show()
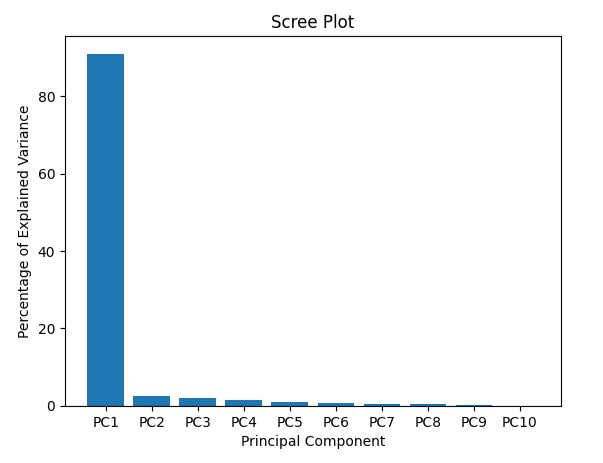
pca_df = pd.DataFrame(pca_data, index=[*wt, *ko], columns=labels)

plt.scatter(pca_df.PC1, pca_df.PC2) plt.title('My PCA Graph') plt.xlabel('PC1 - {}%'.format(per_var[0])) plt.ylabel('PC2 - {}%'.format(per_var[1])) for sample in pca_df.index: plt.annotate(sample, (pca_df.PC1.loc[sample], pca_df.PC2.loc[sample])) plt.show()
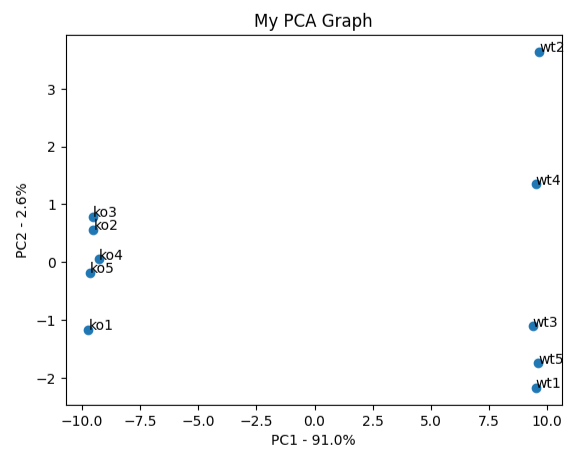
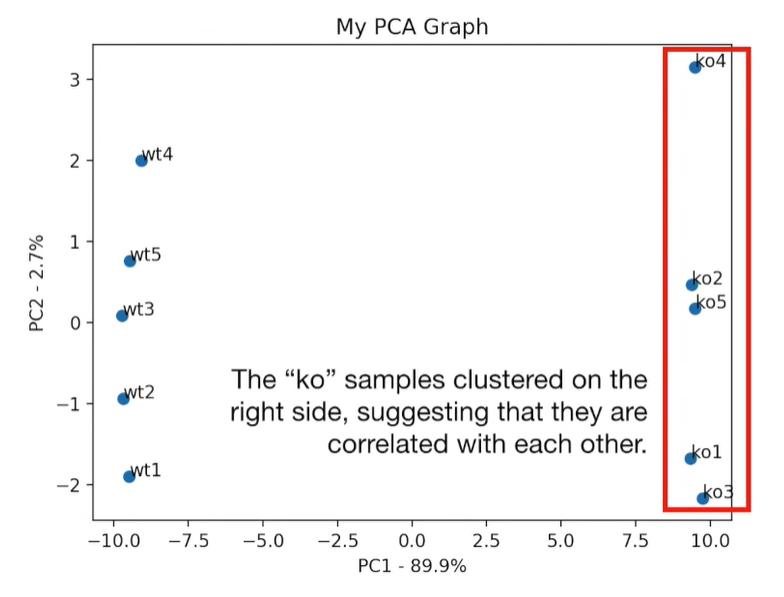
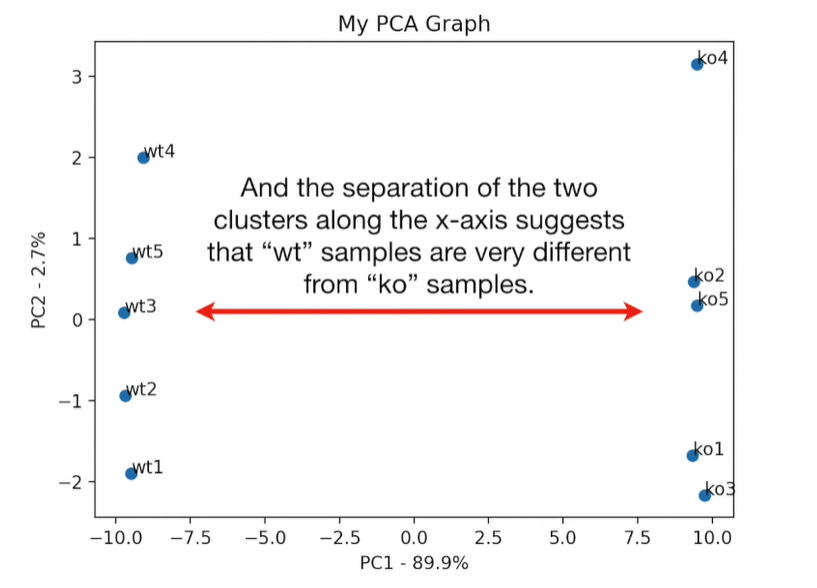

loading_scores = pd.Series(pca.components_[0], index=genes)
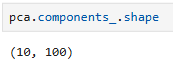

sorted_loading_scores = loading_scores.abs().sort_values(ascending=False)

top_10_genes = sorted_loading_scores[:10].index.values

print(loading_scores[top_10_genes])
gene78 0.104774 gene66 0.104751 gene57 -0.104749 gene16 -0.104740 gene69 -0.104737 gene96 0.104724 gene63 0.104712 gene24 -0.104711 gene81 -0.104699 gene95 -0.104689 dtype: float64





 浙公网安备 33010602011771号
浙公网安备 33010602011771号