ComplexHeatmap 绘制肿瘤突变分布图
来源:https://jokergoo.github.io/ComplexHeatmap-reference/book/
安装
library(devtools)
install_github("jokergoo/ComplexHeatmap")
7.2 Apply to cBioPortal dataset
通过使用实际数据展示oncoPring()的优势。数据来源于cBioPortal。执行步骤如下:
- 登入 http://www.cbioportal.org,
- 查找研究 “Lung Adenocarcinoma Carcinoma” 并选择: “Lung Adenocarcinoma Carcinoma (TCGA, Provisinal)”,
- 在
Enter Gene Set处选择 “General: Ras-Raf-MEK-Erk/JNK signaling (26 genes)”, - 提交
在结果页面, - 点击
Download键,在“Type of Genetic alterations across all cases”处下载text。
样本也可以从结果页面下载, - 点击“OncoPrint”, 移动鼠标到图上,点击“download” 图标,点击“Sample order”.
这组数据已经在ComplexHeatmap包中。首先读入数据,进行前期处理。
mat = read.table(system.file("extdata", package = "ComplexHeatmap",
"tcga_lung_adenocarcinoma_provisional_ras_raf_mek_jnk_signalling.txt"),
header = TRUE, stringsAsFactors = FALSE, sep = "\t")
mat[is.na(mat)] = ""
rownames(mat) = mat[, 1]
mat = mat[, -1]
mat= mat[, -ncol(mat)]
mat = t(as.matrix(mat))
mat[1:3, 1:3]
## TCGA-05-4384-01 TCGA-05-4390-01 TCGA-05-4425-01
## KRAS " " "MUT;" " "
## HRAS " " " " " "
## BRAF " " " " " "
在mat中有三种不同的异常(alternation,不讨论翻译是否准确):HOMDEL,AMP,MUT。首先定义如何添加这些不同的异常。
col = c("HOMDEL" = "blue", "AMP" = "red", "MUT" = "#008000")
alter_fun = list(
background = function(x, y, w, h) {
grid.rect(x, y, w-unit(0.5, "mm"), h-unit(0.5, "mm"),
gp = gpar(fill = "#CCCCCC", col = NA))
},
# big blue
HOMDEL = function(x, y, w, h) {
grid.rect(x, y, w-unit(0.5, "mm"), h-unit(0.5, "mm"),
gp = gpar(fill = col["HOMDEL"], col = NA))
},
# bug red
AMP = function(x, y, w, h) {
grid.rect(x, y, w-unit(0.5, "mm"), h-unit(0.5, "mm"),
gp = gpar(fill = col["AMP"], col = NA))
},
# small green
MUT = function(x, y, w, h) {
grid.rect(x, y, w-unit(0.5, "mm"), h*0.33,
gp = gpar(fill = col["MUT"], col = NA))
}
)
现在,使用oncoPrint。将column_title和heatmap_legend_param保存成向量,因为随后它们会被用到多次。
column_title = "OncoPrint for TCGA Lung Adenocarcinoma, genes in Ras Raf MEK JNK signalling"
heatmap_legend_param = list(title = "Alternations", at = c("HOMDEL", "AMP", "MUT"),
labels = c("Deep deletion", "Amplification", "Mutation"))
oncoPrint(mat,
alter_fun = alter_fun, col = col,
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
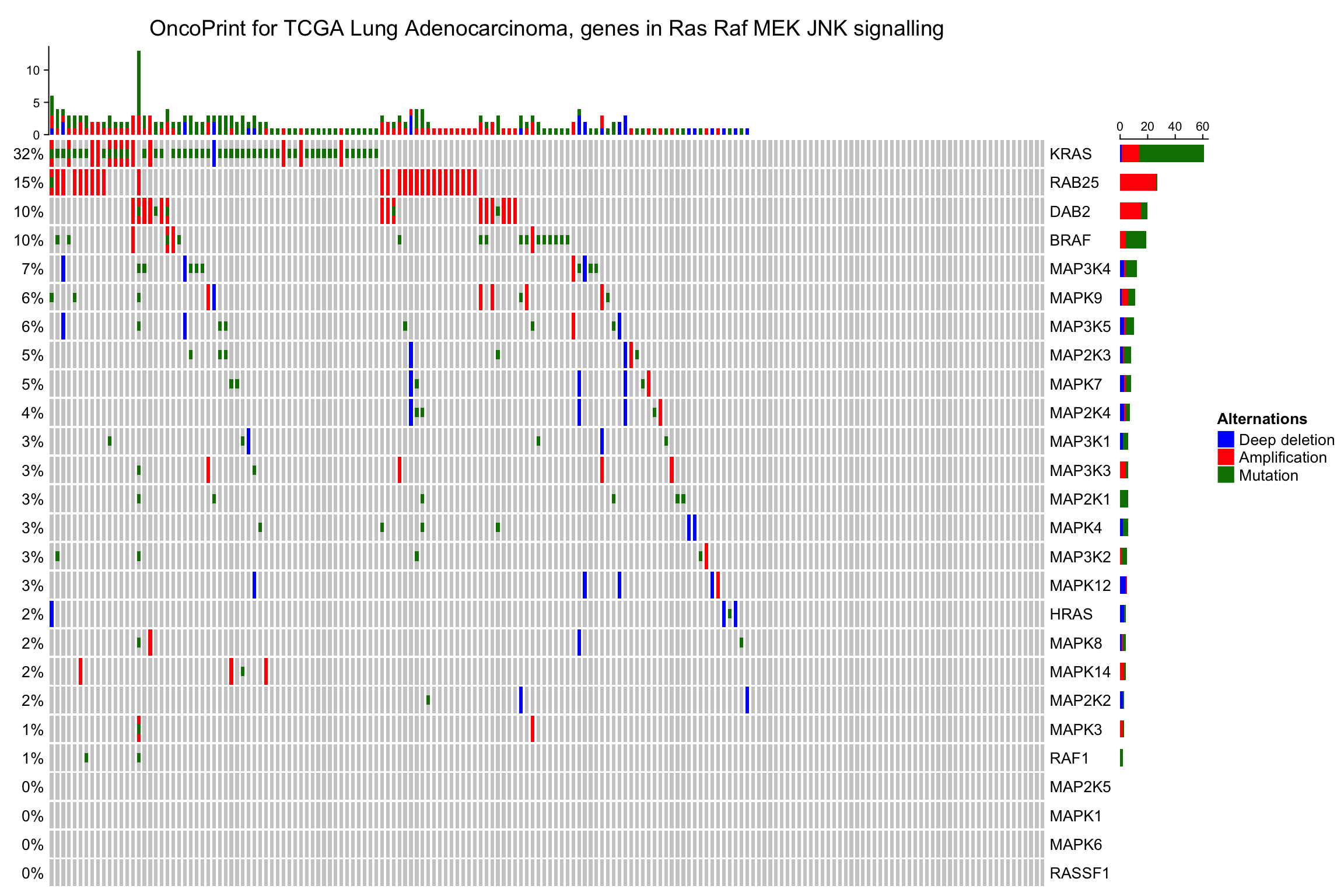
7.2.1 移除空行空列
默认情况下,即使样本没有alteration,仍在热图中,但可以通过设置remove_empty_columns和remove_empty_rows到TRUE来移除这些没有alteration的样本:
oncoPrint(mat,
alter_fun = alter_fun, col = col,
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
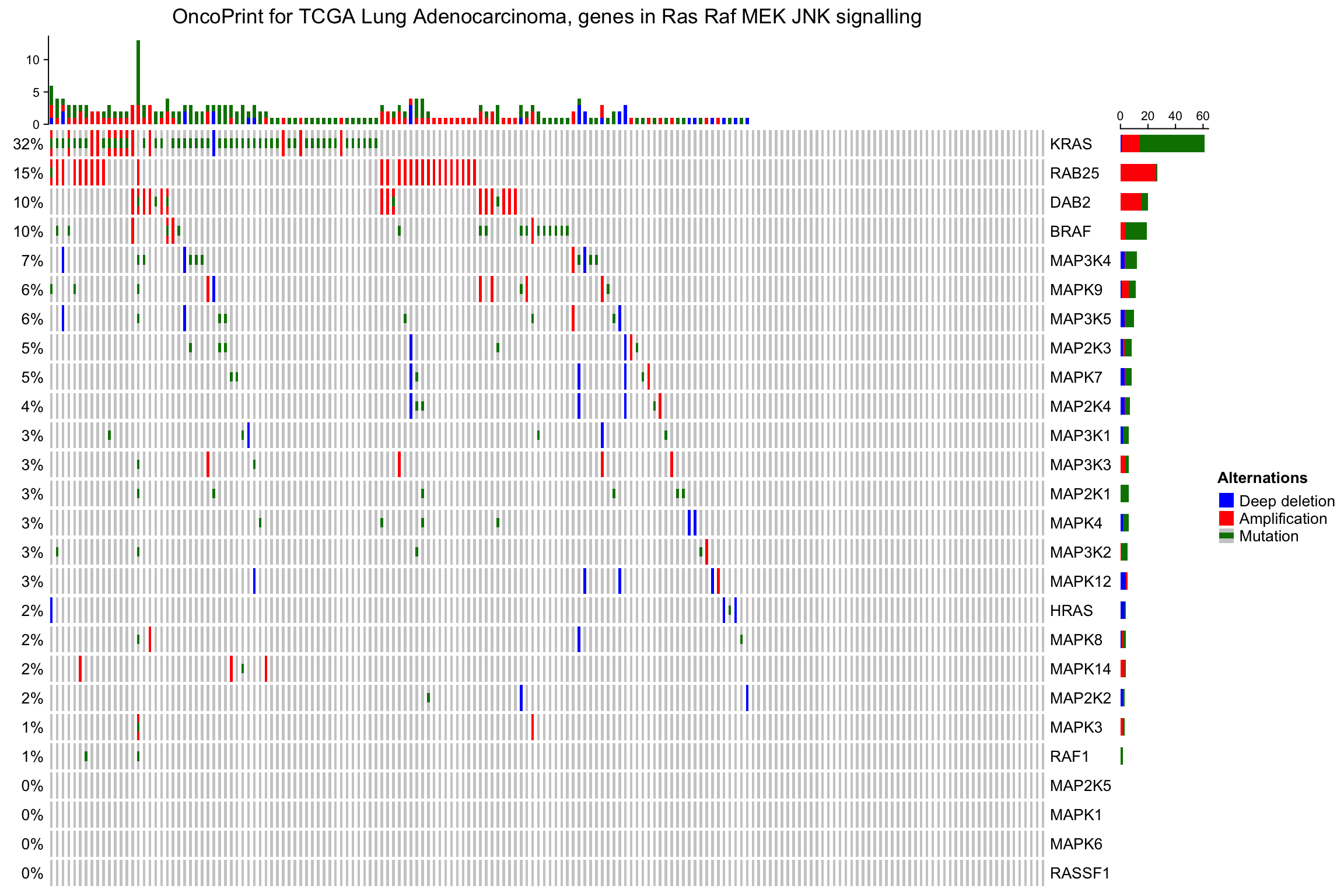
7.2.2 重新排序
同Heatmap()函数已有,row_order或者column_order也能够排序(数字或者字符都行)。以下示例中,样本的顺序来自于cBio。可以看到memo sort和使用cBio的差异。
sample_order = scan(paste0(system.file("extdata", package = "ComplexHeatmap"),
"/sample_order.txt"), what = "character")
oncoPrint(mat,
alter_fun = alter_fun, col = col,
row_order = 1:nrow(mat), column_order = sample_order,
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
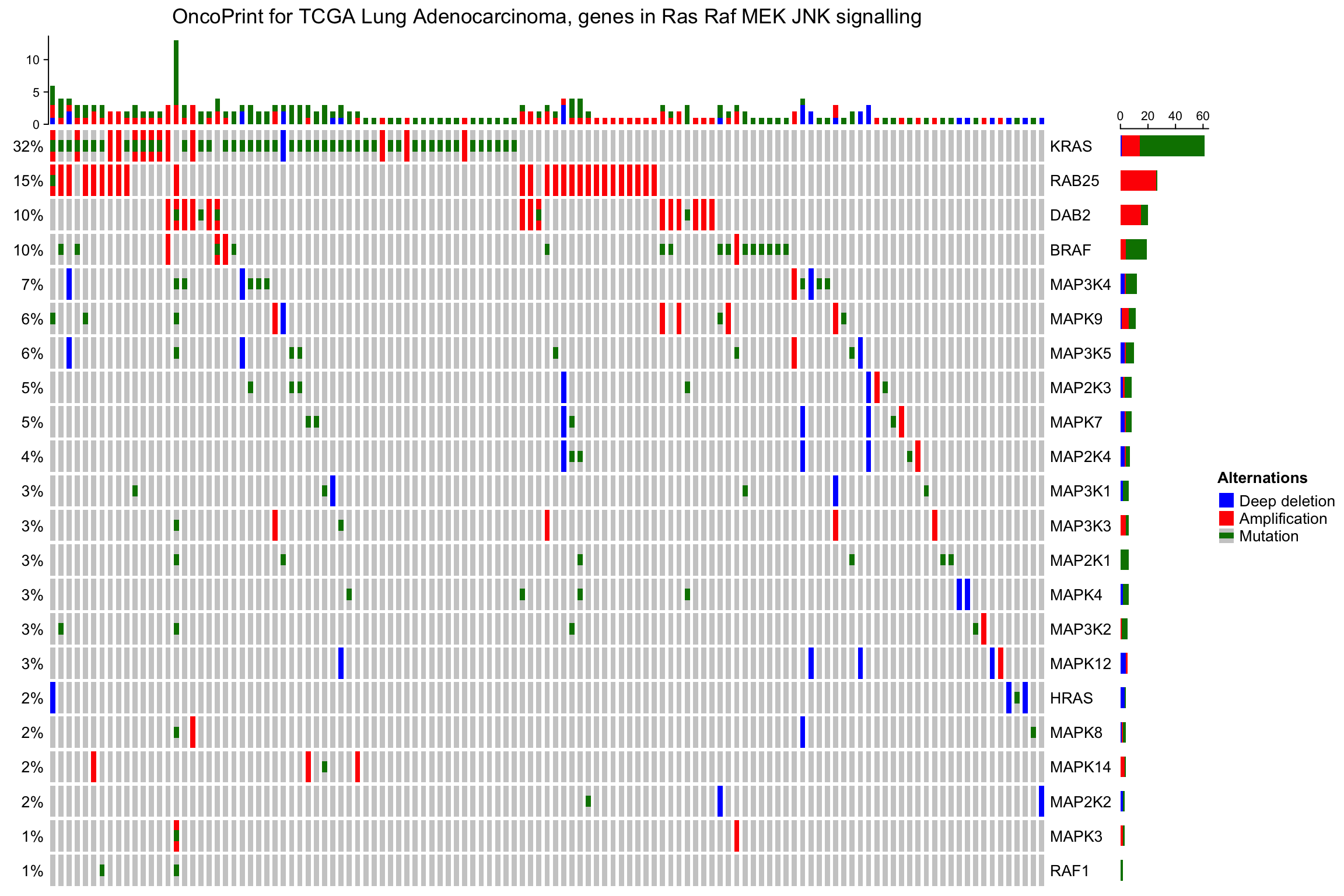
7.2.3 OncoPrint注释
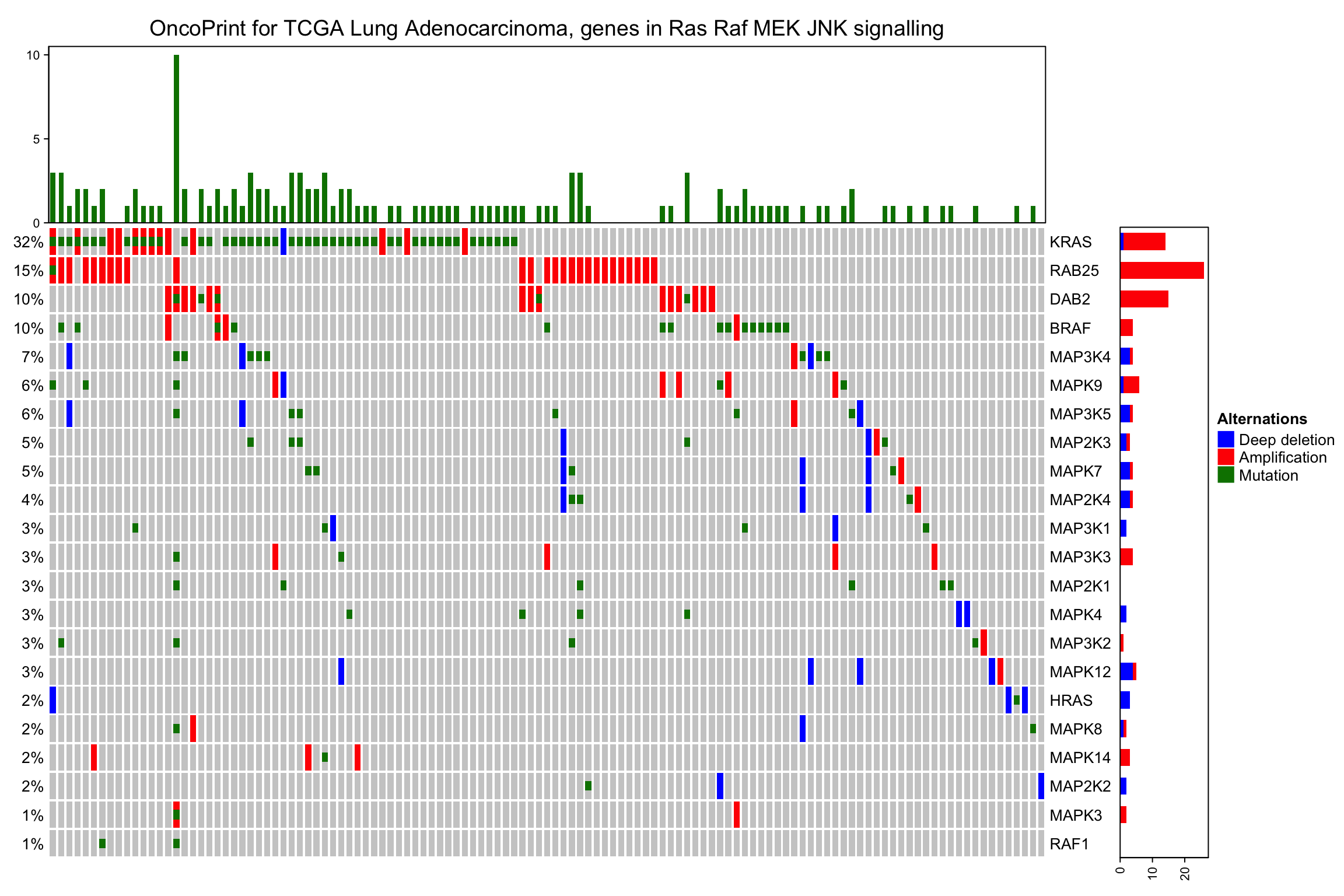
在顶部和右侧,有barplots展示每个基因或者样本的数目,在左侧,展示了每个基因发生突变所在样本的百分比。
barplot注释通过anno_oncoprint_barplot()实现。Barplots默认绘制所有的alteration类型,但是可以选择相应的子集添加到anno_oncoprint_barplot展示。以下是示例:
oncoPrint(mat,
alter_fun = alter_fun, col = col,
top_annotation = HeatmapAnnotation(
column_barplot = anno_oncoprint_barplot("MUT", border = TRUE, # only MUT
height = unit(4, "cm"))),
right_annotation = rowAnnotation(
row_barplot = anno_oncoprint_barplot(c("AMP", "HOMDEL"), # only AMP and HOMDEL
border = TRUE, height = unit(4, "cm"),
axis_param = list(side = "bottom", labels_rot = 90))),
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
可以通过show_pct和show_row_names打开和关闭百分数及行名称。
oncoPrint(mat,
alter_fun = alter_fun, col = col,
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
pct_side = "right", row_names_side = "left",
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
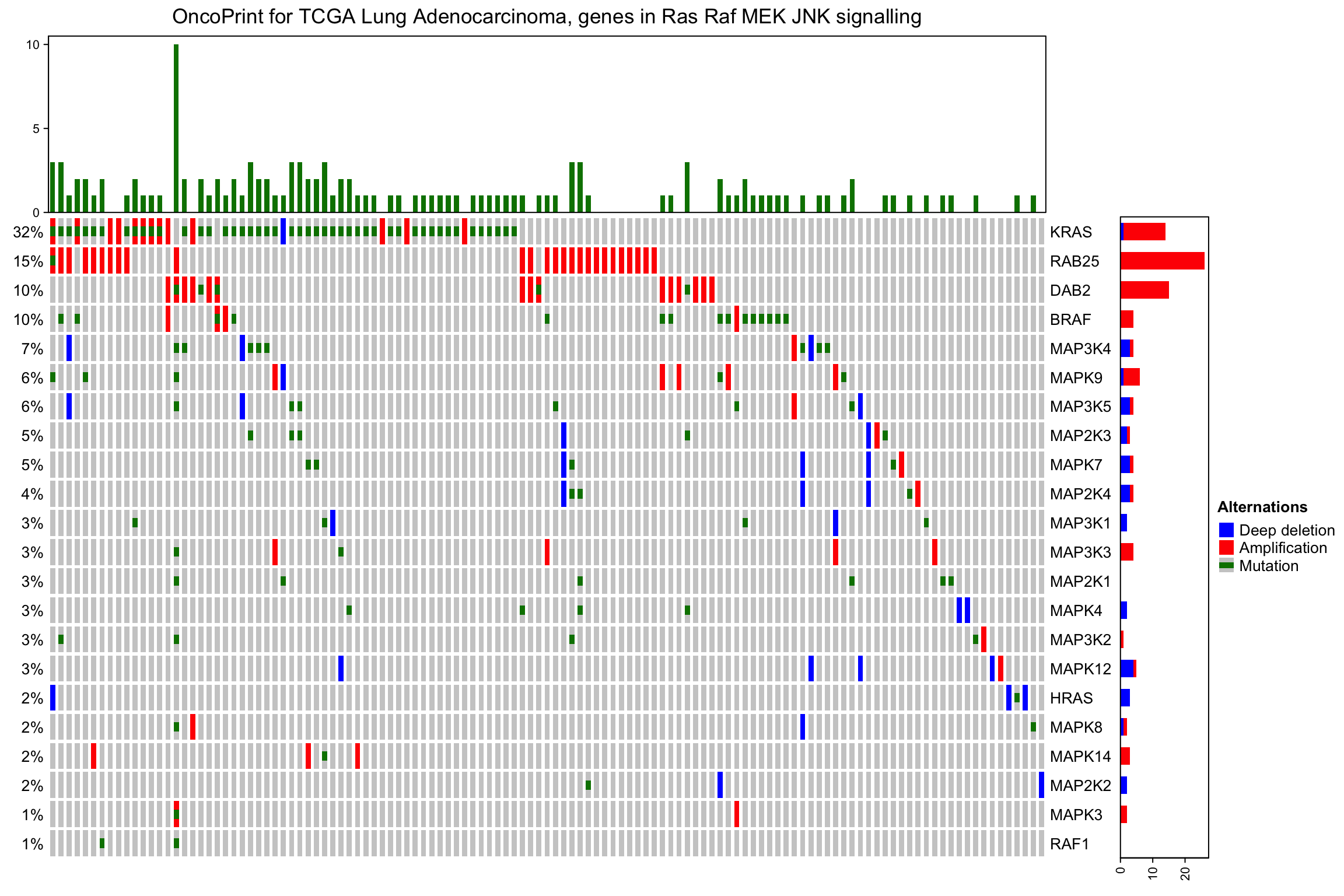
barplot注释属于常规注释,还可以添在HeatmapAnnotation()或者rowAnnotation中加更多的注释:
oncoPrint(mat,
alter_fun = alter_fun, col = col,
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
top_annotation = HeatmapAnnotation(cbar = anno_oncoprint_barplot(),
foo1 = 1:172,
bar1 = anno_points(1:172)),
left_annotation = rowAnnotation(foo2 = 1:26),
right_annotation = rowAnnotation(bar2 = anno_barplot(1:26)),
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
7.2.4 oncoPrint作为热图
oncoPrint()实际上返回一个Heatmap()对象,因此可以水平或者垂直的添加更多的热图和注释,从而时间更加的复杂的展示。
以下示例添加了一个水平方向的热图。
ht_list = oncoPrint(mat,
alter_fun = alter_fun, col = col,
column_title = column_title, heatmap_legend_param = heatmap_legend_param) +
Heatmap(matrix(rnorm(nrow(mat)*10), ncol = 10), name = "expr", width = unit(4, "cm"))
draw(ht_list)
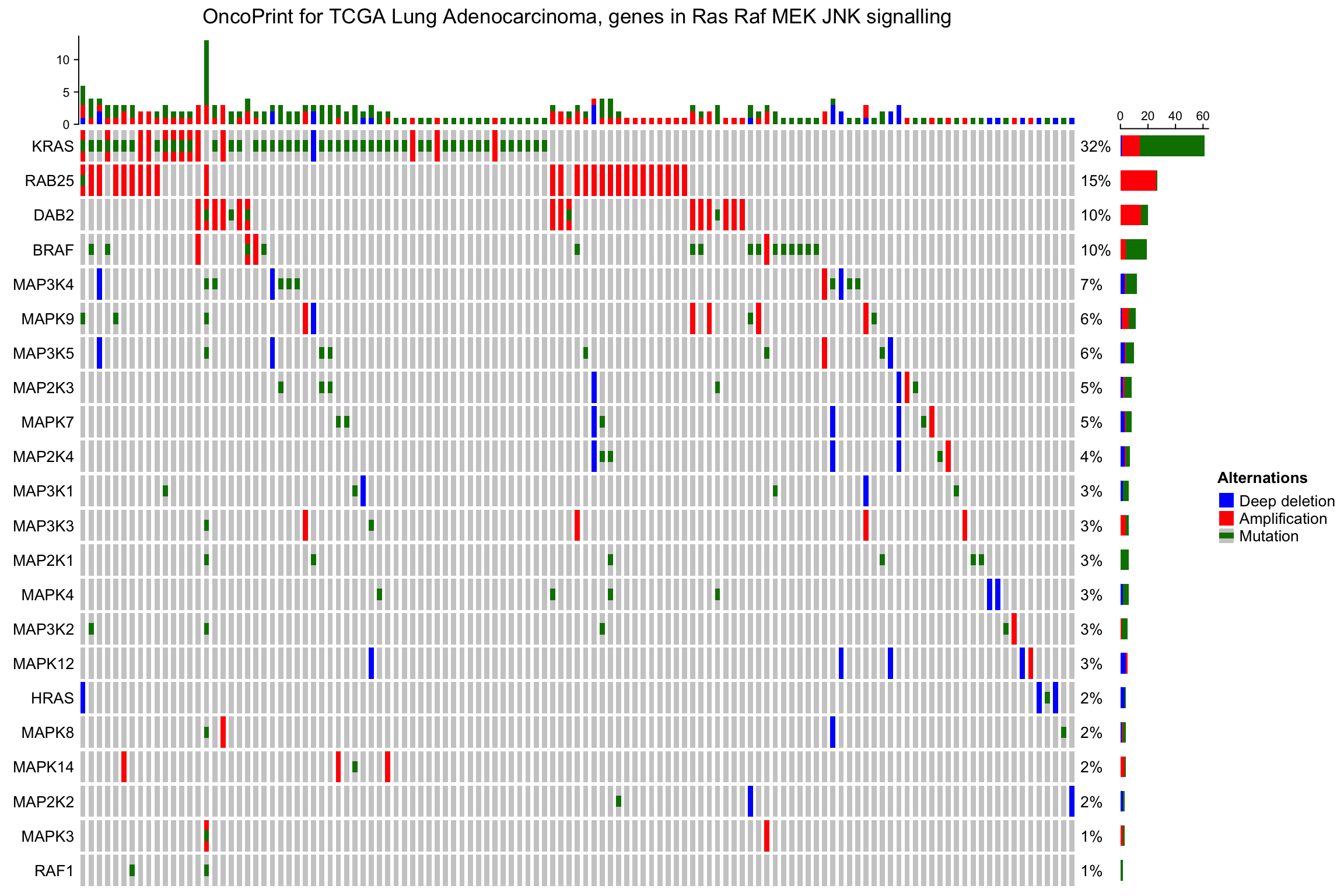
亦可垂直添加:
ht_list = oncoPrint(mat,
alter_fun = alter_fun, col = col,
column_title = column_title, heatmap_legend_param = heatmap_legend_param) %v%
Heatmap(matrix(rnorm(ncol(mat)*10), nrow = 10), name = "expr", height = unit(4, "cm"))
draw(ht_list)
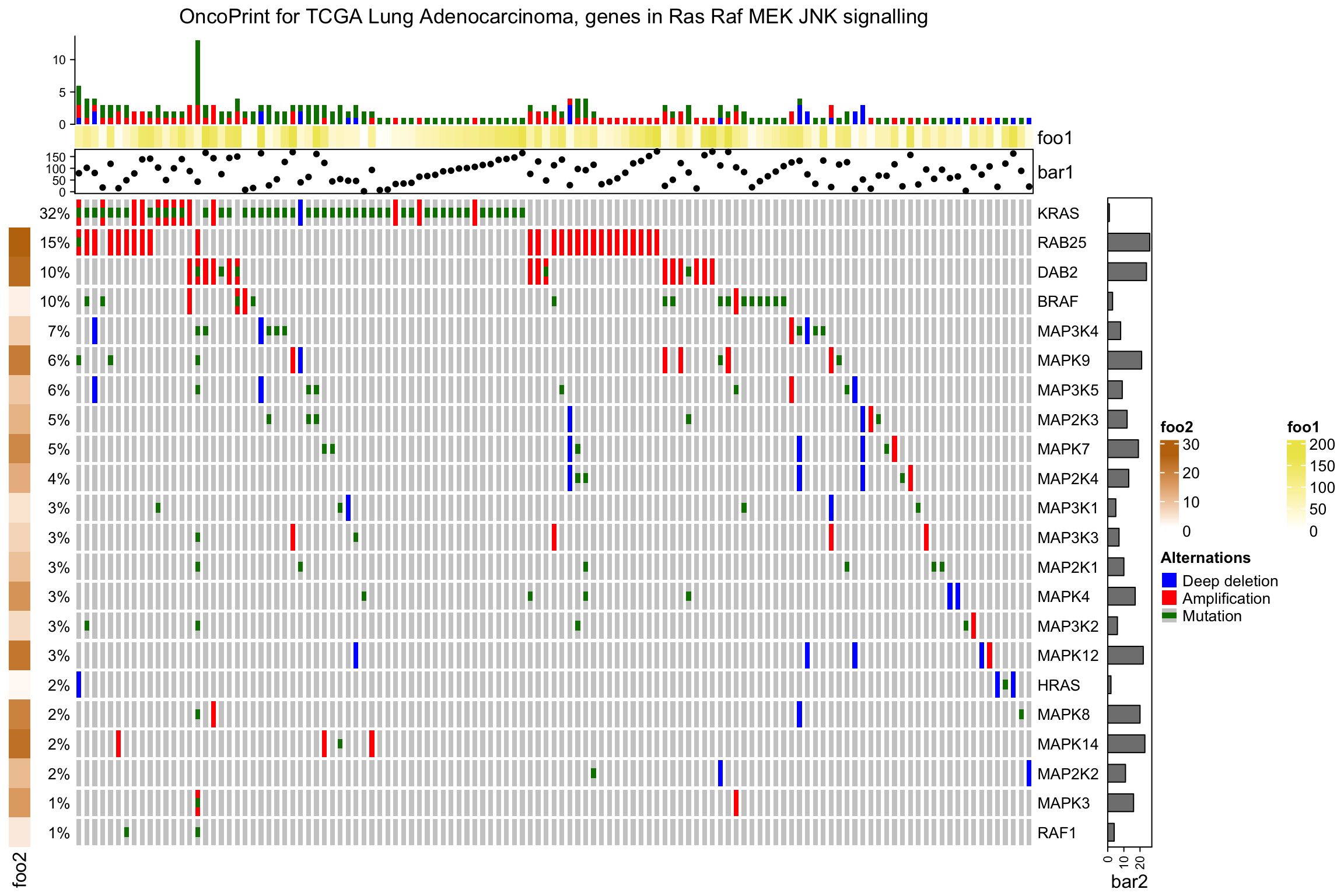
像一般的热图已有,也可以分割热图:
ht_list = oncoPrint(mat,
alter_fun = alter_fun, col = col,
column_title = column_title, heatmap_legend_param = heatmap_legend_param) +
Heatmap(matrix(rnorm(nrow(mat)*10), ncol = 10), name = "expr", width = unit(4, "cm"))
draw(ht_list, row_split = sample(c("a", "b"), nrow(mat), replace = TRUE))
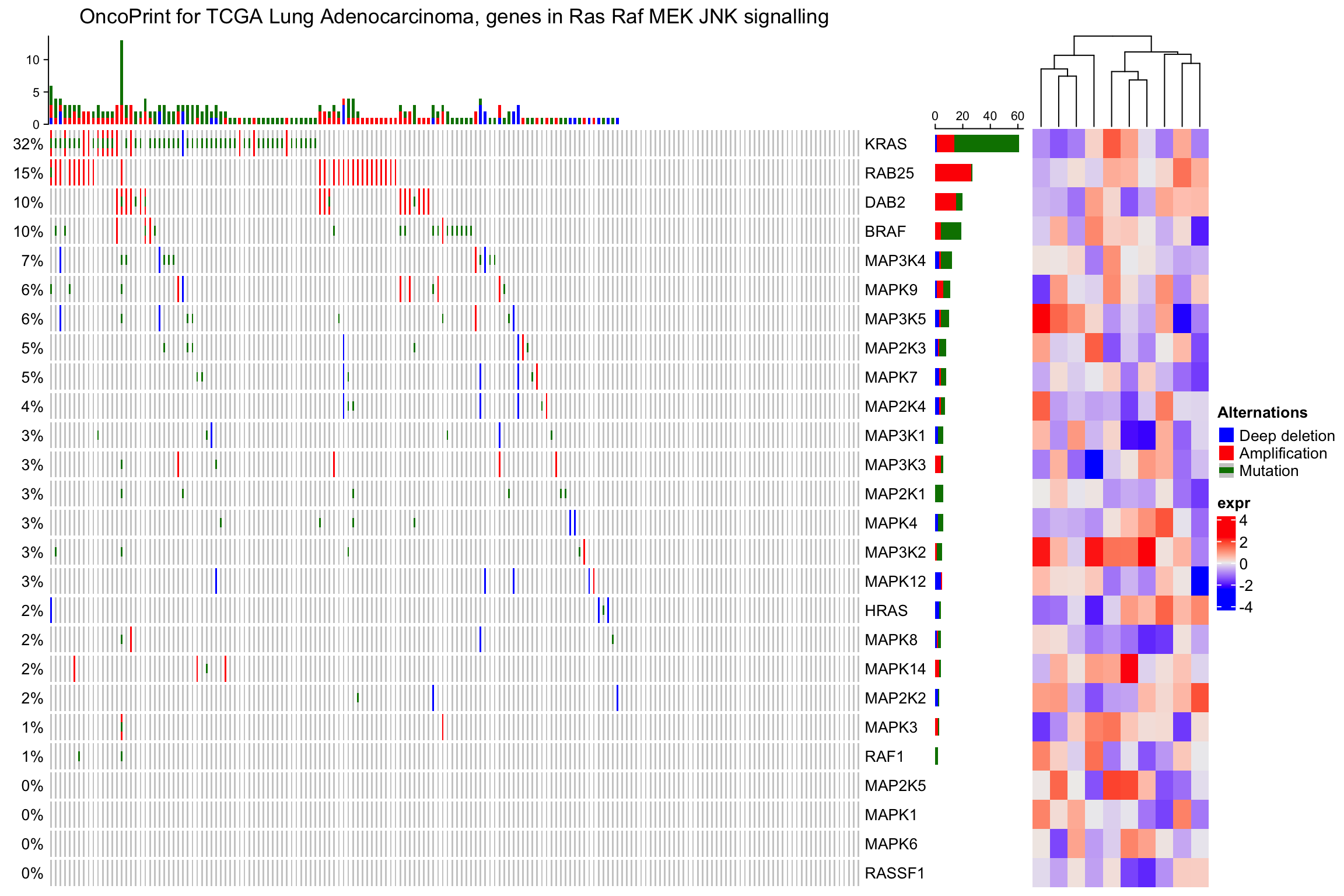
当remove_empty_columns或者remove_empty_rows是设置为TRUE时,基因的数目和样本的数目可能不是原始的。如果原始的矩阵有行名和列名,可通过下面的方式获得:
ht = oncoPrint(mat,
alter_fun = alter_fun, col = col,
remove_empty_columns = TRUE, remove_empty_rows = TRUE,
column_title = column_title, heatmap_legend_param = heatmap_legend_param)
rownames(ht@matrix)
## [1] "KRAS" "HRAS" "BRAF" "RAF1" "MAP3K1" "MAP3K2" "MAP3K3"
## [8] "MAP3K4" "MAP3K5" "MAP2K1" "MAP2K2" "MAP2K3" "MAP2K4" "MAPK3"
## [15] "MAPK4" "MAPK7" "MAPK8" "MAPK9" "MAPK12" "MAPK14" "DAB2"
## [22] "RAB25"
colnames(ht@matrix)
## [1] "TCGA-05-4390-01" "TCGA-38-4631-01" "TCGA-44-6144-01"
## [4] "TCGA-44-6145-01" "TCGA-44-6146-01" "TCGA-49-4488-01"
## [7] "TCGA-50-5930-01" "TCGA-50-5931-01" "TCGA-50-5932-01"
## [10] "TCGA-50-5933-01" "TCGA-50-5941-01" "TCGA-50-5942-01"
## [13] "TCGA-50-5944-01" "TCGA-50-5946-01" "TCGA-50-6591-01"
## [16] "TCGA-50-6592-01" "TCGA-50-6594-01" "TCGA-67-4679-01"
## [19] "TCGA-67-6215-01" "TCGA-73-4658-01" "TCGA-73-4676-01"
## [22] "TCGA-75-5122-01" "TCGA-75-5125-01" "TCGA-75-5126-01"
## [25] "TCGA-75-6206-01" "TCGA-75-6211-01" "TCGA-86-6562-01"
## [28] "TCGA-05-4396-01" "TCGA-05-4405-01" "TCGA-05-4410-01"
## [31] "TCGA-05-4415-01" "TCGA-05-4417-01" "TCGA-05-4424-01"
## [34] "TCGA-05-4427-01" "TCGA-05-4433-01" "TCGA-44-6774-01"
## [37] "TCGA-44-6775-01" "TCGA-44-6776-01" "TCGA-44-6777-01"
## [40] "TCGA-44-6778-01" "TCGA-49-4487-01" "TCGA-49-4490-01"
## [43] "TCGA-49-6744-01" "TCGA-49-6745-01" "TCGA-49-6767-01"
## [46] "TCGA-50-5044-01" "TCGA-50-5051-01" "TCGA-50-5072-01"
## [49] "TCGA-50-6590-01" "TCGA-55-6642-01" "TCGA-55-6712-01"
## [52] "TCGA-71-6725-01" "TCGA-91-6828-01" "TCGA-91-6829-01"
## [55] "TCGA-91-6835-01" "TCGA-91-6836-01" "TCGA-35-3615-01"
## [58] "TCGA-44-2655-01" "TCGA-44-2656-01" "TCGA-44-2662-01"
## [61] "TCGA-44-2666-01" "TCGA-44-2668-01" "TCGA-55-1592-01"
## [64] "TCGA-55-1594-01" "TCGA-55-1595-01" "TCGA-64-1676-01"
## [67] "TCGA-64-1677-01" "TCGA-64-1678-01" "TCGA-64-1680-01"
## [70] "TCGA-67-3771-01" "TCGA-67-3773-01" "TCGA-67-3774-01"
## [73] "TCGA-05-4244-01" "TCGA-05-4249-01" "TCGA-05-4250-01"
## [76] "TCGA-35-4122-01" "TCGA-35-4123-01" "TCGA-44-2657-01"
## [79] "TCGA-44-3398-01" "TCGA-44-3918-01" "TCGA-05-4382-01"
## [82] "TCGA-05-4389-01" "TCGA-05-4395-01" "TCGA-05-4397-01"
## [85] "TCGA-05-4398-01" "TCGA-05-4402-01" "TCGA-05-4403-01"
## [88] "TCGA-05-4418-01" "TCGA-05-4420-01" "TCGA-05-4422-01"
## [91] "TCGA-05-4426-01" "TCGA-05-4430-01" "TCGA-05-4434-01"
## [94] "TCGA-38-4625-01" "TCGA-38-4626-01" "TCGA-38-4628-01"
## [97] "TCGA-38-4630-01" "TCGA-44-3396-01" "TCGA-49-4486-01"
## [100] "TCGA-49-4505-01" "TCGA-49-4506-01" "TCGA-49-4507-01"
## [103] "TCGA-49-4510-01" "TCGA-73-4659-01" "TCGA-73-4662-01"
## [106] "TCGA-73-4668-01" "TCGA-73-4670-01" "TCGA-73-4677-01"
## [109] "TCGA-05-5428-01" "TCGA-05-5715-01" "TCGA-50-5045-01"
## [112] "TCGA-50-5049-01" "TCGA-50-5936-01" "TCGA-55-5899-01"
## [115] "TCGA-64-5774-01" "TCGA-64-5775-01" "TCGA-64-5778-01"
## [118] "TCGA-64-5815-01" "TCGA-75-5146-01" "TCGA-75-5147-01"
## [121] "TCGA-80-5611-01"




 浙公网安备 33010602011771号
浙公网安备 33010602011771号