![image]()
绘图代码
import torch
import copy
import matplotlib.pyplot as plt
import numpy as np
from matplotlib.patches import Rectangle
# 存储每个蛋白质的结果
protein_results = []
with torch.no_grad():
for row, data in zip(rows, test_graphs):
if len(row[2]) >= 36:
name = row[0]
data = copy.deepcopy(data)
data = data.cuda()
out, wp = model(data)
indices = wp[0].mean(0).cpu().numpy().argsort()[:][::-1]
num = int(len(indices) * 0.1)
indices = indices[:num]
# 确保所有位点都是整数类型
true_sites = [int(x) for x in row[2]]
intersection = list(set(indices) & set(true_sites))
ratio = len(intersection) / len(true_sites) if len(true_sites) > 0 else 0
protein_results.append({
'name': name,
'true_sites': true_sites,
'predicted_sites': indices,
'intersection': intersection,
'ratio': ratio
})
# 按重合率排序并选择前10个
protein_results.sort(key=lambda x: x['ratio'], reverse=True)
top_proteins = protein_results[:6] # 取前5个
# 如果没有符合条件的蛋白质,显示提示信息
if not top_proteins:
print("No proteins with at least 36 sites found.")
exit()
# 创建可视化 - 每个蛋白质一行
fig, axes = plt.subplots(len(top_proteins), 1, figsize=(15, 1.5 * len(top_proteins)))
# 如果只有一个蛋白质,确保axes是列表形式
if len(top_proteins) == 1:
axes = [axes]
# 减小子图之间的间距
plt.subplots_adjust(hspace=-0.9) # 减小垂直间距
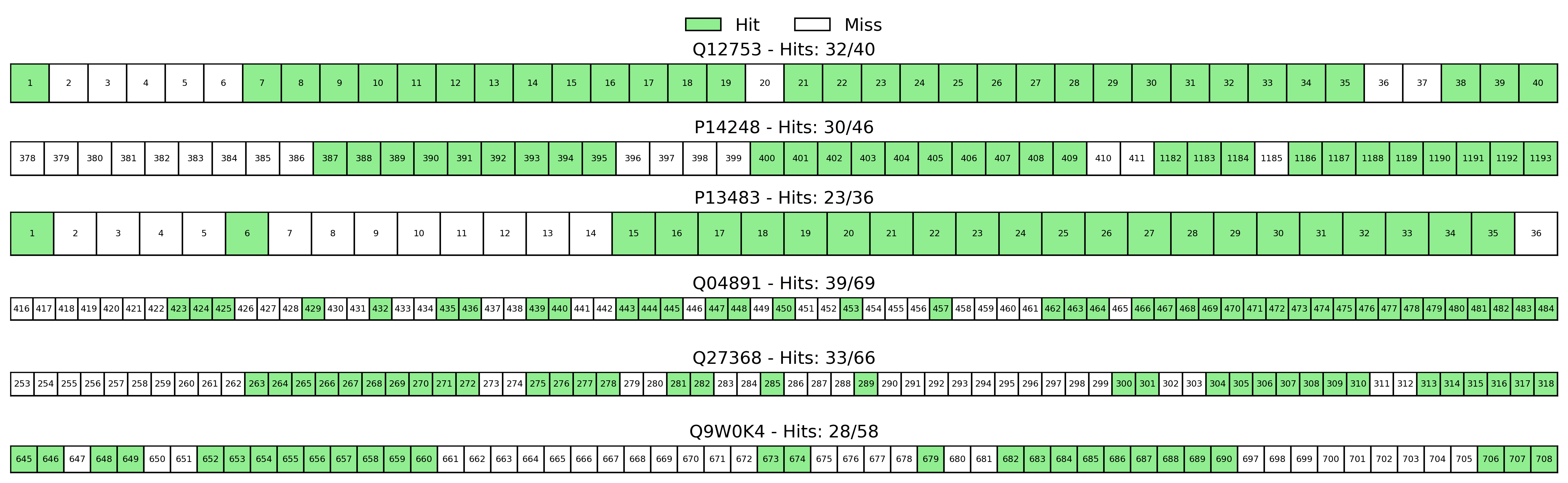
for i, (ax, protein) in enumerate(zip(axes, top_proteins)):
name = protein['name']
true_sites = sorted(protein['true_sites'])
intersection = protein['intersection']
# 绘制一行格子
for j, site in enumerate(true_sites):
# 确定颜色:如果在交集中则为绿色,否则为无色
is_hit = site in intersection
color = 'lightgreen' if is_hit else 'none'
rect = Rectangle((j, 0), 1, 1, facecolor=color, edgecolor='black')
ax.add_patch(rect)
# 添加文本标签
ax.text(j + 0.5, 0.5, str(site),
ha='center', va='center', fontsize=6)
# 设置图表属性
ax.set_xlim(0, len(true_sites))
ax.set_ylim(0, 1)
ax.set_aspect('equal')
# 修改标题:显示具体的击中数量/总位点
ax.set_title(f'{name} - Hits: {len(intersection)}/{len(true_sites)}')
ax.set_xticks([])
ax.set_yticks([])
# # 添加图例 - 放在最上面
legend_elements = [
plt.Rectangle((0,0),1,1, facecolor='lightgreen', edgecolor='black', label='Hit'),
plt.Rectangle((0,0),1,1, facecolor='none', edgecolor='black', label='Miss')
]
fig.legend(handles=legend_elements, loc='upper center', bbox_to_anchor=(0.5, 0.76), # 调整位置 bbox_to_anchor=(0.5, 0.85
ncol=2, frameon=False, fontsize=12)
plt.tight_layout(rect=[0, 0, 1, 0.95]) # 调整布局,为顶部图例留出空间
# 保存为300dpi的矢量图
plt.savefig('protein_hits_visualization.pdf', dpi=300, bbox_inches='tight')
plt.savefig('protein_hits_visualization.png', dpi=300, bbox_inches='tight')
plt.show()
# 打印统计结果
ans = sum(1 for p in protein_results if len(p['intersection']) > 0)
sums = len(protein_results)
print(f"Detection Rate: {ans}/{sums} = {ans/sums:.4f}")
# 打印前5个蛋白质的详细信息
print("\nTop proteins by overlap ratio (≥36 sites):")
for i, protein in enumerate(top_proteins, 1):
print(f"{i}. {protein['name']}: Hits = {len(protein['intersection'])}/{len(protein['true_sites'])}" +
f" (Ratio = {protein['ratio']:.4f})")



 浙公网安备 33010602011771号
浙公网安备 33010602011771号