









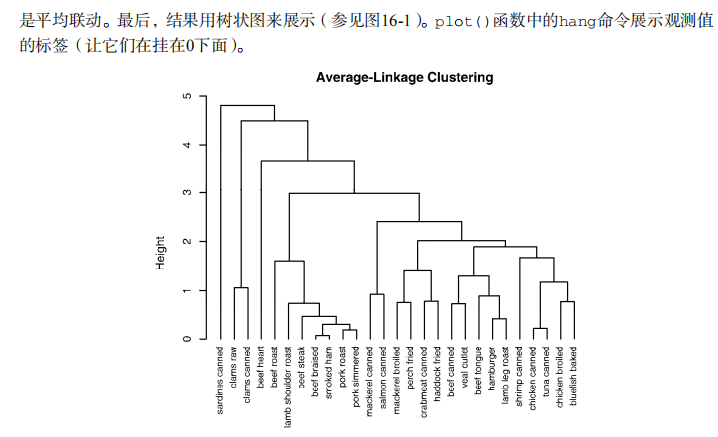


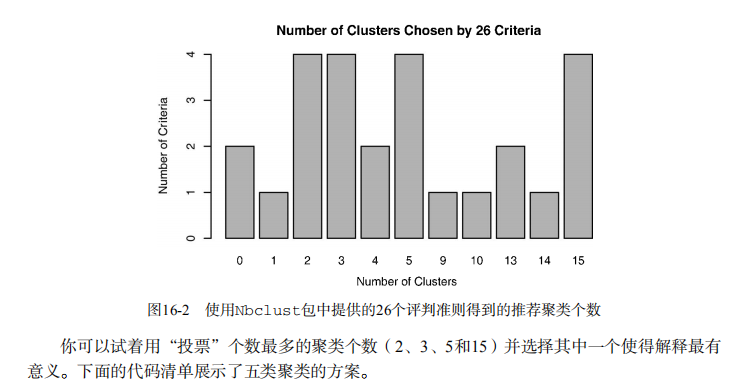

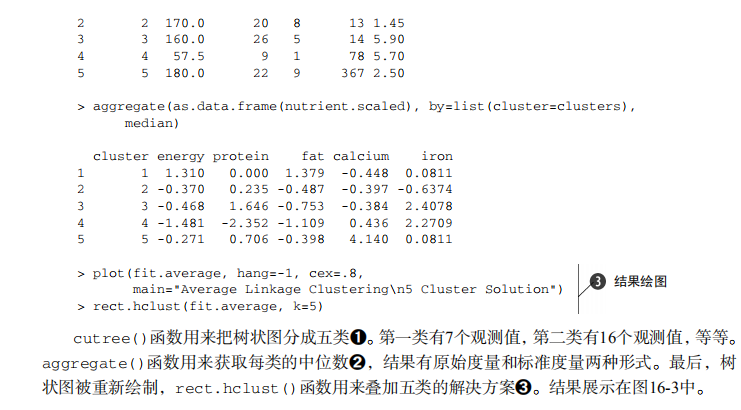
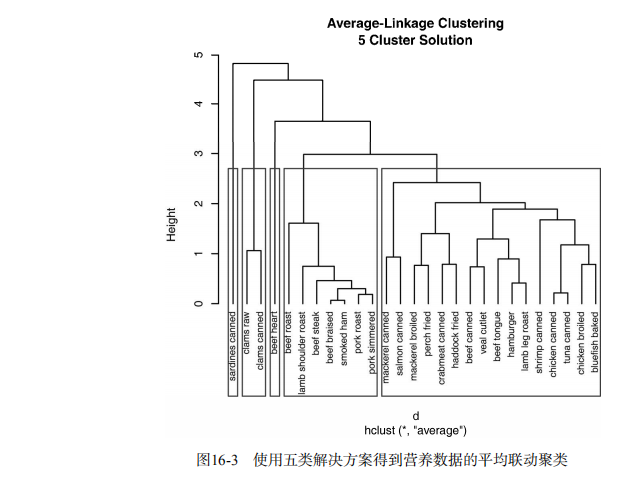

#-------------------------------------------------------# # R in Action (2nd ed): Chapter 16 # # Cluster analysis # # requires packaged NbClust, flexclust, rattle # # install.packages(c("NbClust", "flexclust", "rattle")) # #-------------------------------------------------------# par(ask=TRUE) opar <- par(no.readonly=FALSE) # Calculating Distances data(nutrient, package="flexclust") head(nutrient, 2) d <- dist(nutrient) as.matrix(d)[1:4,1:4] # Listing 16.1 - Average linkage clustering of nutrient data data(nutrient, package="flexclust") row.names(nutrient) <- tolower(row.names(nutrient)) nutrient.scaled <- scale(nutrient) d <- dist(nutrient.scaled) fit.average <- hclust(d, method="average") plot(fit.average, hang=-1, cex=.8, main="Average Linkage Clustering") # Listing 16.2 - Selecting the number of clusters library(NbClust) nc <- NbClust(nutrient.scaled, distance="euclidean", min.nc=2, max.nc=15, method="average") par(opar) table(nc$Best.n[1,]) barplot(table(nc$Best.n[1,]), xlab="Numer of Clusters", ylab="Number of Criteria", main="Number of Clusters Chosen by 26 Criteria") # Listing 16.3 - Obtaining the final cluster solution clusters <- cutree(fit.average, k=5) table(clusters) aggregate(nutrient, by=list(cluster=clusters), median) aggregate(as.data.frame(nutrient.scaled), by=list(cluster=clusters), median) plot(fit.average, hang=-1, cex=.8, main="Average Linkage Clustering\n5 Cluster Solution") rect.hclust(fit.average, k=5) # Plot function for within groups sum of squares by number of clusters wssplot <- function(data, nc=15, seed=1234){ wss <- (nrow(data)-1)*sum(apply(data,2,var)) for (i in 2:nc){ set.seed(seed) wss[i] <- sum(kmeans(data, centers=i)$withinss)} plot(1:nc, wss, type="b", xlab="Number of Clusters", ylab="Within groups sum of squares")} # Listing 16.4 - K-means clustering of wine data data(wine, package="rattle") head(wine) df <- scale(wine[-1]) wssplot(df) library(NbClust) set.seed(1234) nc <- NbClust(df, min.nc=2, max.nc=15, method="kmeans") par(opar) table(nc$Best.n[1,]) barplot(table(nc$Best.n[1,]), xlab="Numer of Clusters", ylab="Number of Criteria", main="Number of Clusters Chosen by 26 Criteria") set.seed(1234) fit.km <- kmeans(df, 3, nstart=25) fit.km$size fit.km$centers aggregate(wine[-1], by=list(cluster=fit.km$cluster), mean) # evaluate clustering ct.km <- table(wine$Type, fit.km$cluster) ct.km library(flexclust) randIndex(ct.km) # Listing 16.5 - Partitioning around mediods for the wine data library(cluster) set.seed(1234) fit.pam <- pam(wine[-1], k=3, stand=TRUE) fit.pam$medoids clusplot(fit.pam, main="Bivariate Cluster Plot") # evaluate clustering ct.pam <- table(wine$Type, fit.pam$clustering) ct.pam randIndex(ct.pam) ## Avoiding non-existent clusters library(fMultivar) set.seed(1234) df <- rnorm2d(1000, rho=.5) df <- as.data.frame(df) plot(df, main="Bivariate Normal Distribution with rho=0.5") wssplot(df) library(NbClust) nc <- NbClust(df, min.nc=2, max.nc=15, method="kmeans") par(opar) barplot(table(nc$Best.n[1,]), xlab="Numer of Clusters", ylab="Number of Criteria", main ="Number of Clusters Chosen by 26 Criteria") library(ggplot2) library(cluster) fit <- pam(df, k=2) df$clustering <- factor(fit$clustering) ggplot(data=df, aes(x=V1, y=V2, color=clustering, shape=clustering)) + geom_point() + ggtitle("Clustering of Bivariate Normal Data") plot(nc$All.index[,4], type="o", ylab="CCC", xlab="Number of clusters", col="blue")



