单细胞最好的教程(十八): 细胞类型映射到细胞本体论:让你的单细胞注释更专业!
细胞类型映射到细胞本体论:让你的单细胞注释更专业!
作者按
在单细胞数据分析领域,标准化的细胞类型注释对于数据整合和比较研究至关重要。本文将介绍如何使用Cell Ontology(细胞本体论)来规范化你的细胞类型注释,提高研究的可重复性和可比性。本教程首发于单细胞最好的中文教程,未经授权许可,禁止转载。
全文字数|预计阅读时间: ~4000 | 5min
——Starlitnightly(星夜)
🔍 什么是细胞本体论(Cell Ontology)?
细胞本体论(CL)是一个专门用于分类和描述不同生物体中细胞类型的标准化系统。作为模式生物和生物信息学数据库的重要资源,它具有以下特点:
- 📚 包含超过2700种动物细胞类型的详细分类
- 提供高层次的细胞类型分类标准
- 可以作为其他物种(如植物本体论或果蝇解剖学本体论)中细胞类型的映射参考
- 与其他本体论(如Uberon、GO、CHEBI、PR和PATO)无缝集成
- 能够将细胞类型与解剖结构、生物过程等相关概念建立联系
💡 提示:使用标准化的细胞本体论可以大大提高你的研究结果在国际上的认可度和引用率!
细胞本体 (CL) 创建于 2004 年,自 OBO Foundry 成立以来一直是其核心本体。自那时起,CL 已被各种项目采用,包括 HuBMAP 项目、人类细胞图谱 (HCA)、cellxgene 平台、单细胞表达图谱、BRAIN 倡议细胞普查网络 (BICCN)、ArrayExpress、细胞图像库 (The Cell Image Library)、ENCODE 和 FANTOM5,用于注释细胞类型并促进细胞参考图谱绘制
什么是国家生物信息学中心细胞分类学(Cell Taxonomy)
细胞分类学 (Cell Taxonomy) 基于 4,299 篇出版物和约 350 万个细胞的单细胞转录组谱,对细胞类型和细胞标记物进行了多方面的表征,包括细胞标记物和细胞簇的质量评估、跨物种比较、组织细胞组成以及基于标记物的细胞相似性分析。
💡 提示:细胞分类学的优势是其来自官方倡议,并且有更多的文献和marker作为支撑,细胞类型命名更有权威性
不过细胞本体论的更新是一直持续的,细胞分类学自2023年论文发表后,细胞类型好像就一直没更新了,所以通过两个数据库的命名来规范一起做,我觉得是一个不错的选择。
在这里,我们提供了几个强大的函数,可以将你注释的细胞名称智能转换为对应的细胞本体论名称和ID。所有分析都通过omicverse.single.CellOntologyMapper类来完成。让我们开始动手实践吧!
import scanpy as sc
#import pertpy as pt
import omicverse as ov
ov.plot_set()
%load_ext autoreload
%autoreload 2
📊 数据准备
在开始转换细胞名称之前,你需要先完成细胞注释。在本教程中,我们使用了来自pertpy的haber_2017_regions数据集作为示例。这是一个来自小肠的单细胞测序数据集,包含了多种上皮细胞类型。
import pertpy as pt
adata = pt.dt.haber_2017_regions()
adata.obs['cell_label'].unique()
['Enterocyte.Progenitor', 'Stem', 'TA.Early', 'TA', 'Tuft', 'Enterocyte', 'Goblet', 'Endocrine']
Categories (8, object): ['Endocrine', 'Enterocyte', 'Enterocyte.Progenitor', 'Goblet', 'Stem', 'TA', 'TA.Early', 'Tuft']
⬇️ 下载CL模型
在开始分析之前,我们需要从Cell Ontology下载cl.json文件。这个文件包含了完整的细胞本体论数据库。我们提供了多种下载方式:
方式一:命令行下载
# 从OBO页面下载cl.ono
!mkdir new_ontology
!wget http://purl.obolibrary.org/obo/cl/cl.json -O new_ontology/cl.json
方式二:自动下载(推荐新手)
我们提供了一个名为omicverse.single.download_cl()的函数来自动完成下载过程。这个函数特别智能,即使遇到网络问题,它也能自动选择最佳的下载源。
方式三:手动下载(网络受限时的备选方案)
如果你的网络访问受限,可以使用以下链接手动下载:
- 🌏 Google Drive链接:https://drive.google.com/uc?export=download&id=1niokr5INjWFVjiXHfdCoWioh0ZEYCPkv
- 🇨🇳 蓝奏云链接(国内用户推荐):https://www.lanzoup.com/iN6CX2ybh48h
ov.single.download_cl(output_dir="new_ontology", filename="cl.json")
Downloading Cell Ontology to: new_ontology/cl.json
============================================================
[1/3] Trying Official OBO Library...
URL: http://purl.obolibrary.org/obo/cl/cl.json
Description: Direct download from official Cell Ontology
→ Downloading...
🛠️ 配置CellOntologyMapper
CellOntologyMapper的核心是基于SentenceTransformer的NLP嵌入模型。选择合适的模型对于映射效果至关重要:
| 模型名称 | 特点 | 适用场景 |
|---|---|---|
| BAAI/bge-base-en-v1.5 | 性能最优 | 需要高精度的正式分析 |
| BAAI/bge-small-en-v1.5 | 速度快 | 快速测试或小规模数据 |
| sentence-transformers/all-MiniLM-L6-v2 | 平衡型 | 日常分析使用 |
你也可以在huggingface的官网找到更多的模型:https://hf-mirror.com/models?library=sentence-transformers
💡 小贴士:如果你的计算资源充足,建议使用BAAI/bge-base-en-v1.5获得最佳效果。
#
mapper = ov.single.CellOntologyMapper(
cl_obo_file="new_ontology/cl.json",
model_name="sentence-transformers/all-MiniLM-L6-v2",
local_model_dir="./my_models"
)
🔨 Creating ontology resources from OBO file...
📖 Parsing ontology file...
🧠 Creating NLP embeddings...
🔄 Loading model sentence-transformers/all-MiniLM-L6-v2...
🌐 Checking network connectivity...
✓ Network connection available
🇨🇳 Using HF-Mirror (hf-mirror.com) for faster downloads in China
📁 Models will be saved to: ./my_models
🪞 Downloading model from HF-Mirror: sentence-transformers/all-MiniLM-L6-v2
✓ Model loaded successfully from HF-Mirror!
🔄 Encoding 16841 ontology labels...
你也可以直接加载运行计算好的细胞本体的嵌入,这对于cpu用户而言特别有帮助。
mapper = ov.single.CellOntologyMapper(
cl_obo_file="new_ontology/cl.json",
embeddings_path='new_ontology/ontology_embeddings.pkl',
local_model_dir="./my_models"
)
📥 Loading existing ontology embeddings...
📥 Loaded embeddings for 16841 ontology labels
📋 Ontology mappings loaded: 16841 cell types
细胞类型名称映射
我们可以使用 map_adata 来直接映射我们的细胞类型,并且我们可以可视化
mapping_results = mapper.map_adata(
adata,
cell_name_col='cell_label'
)
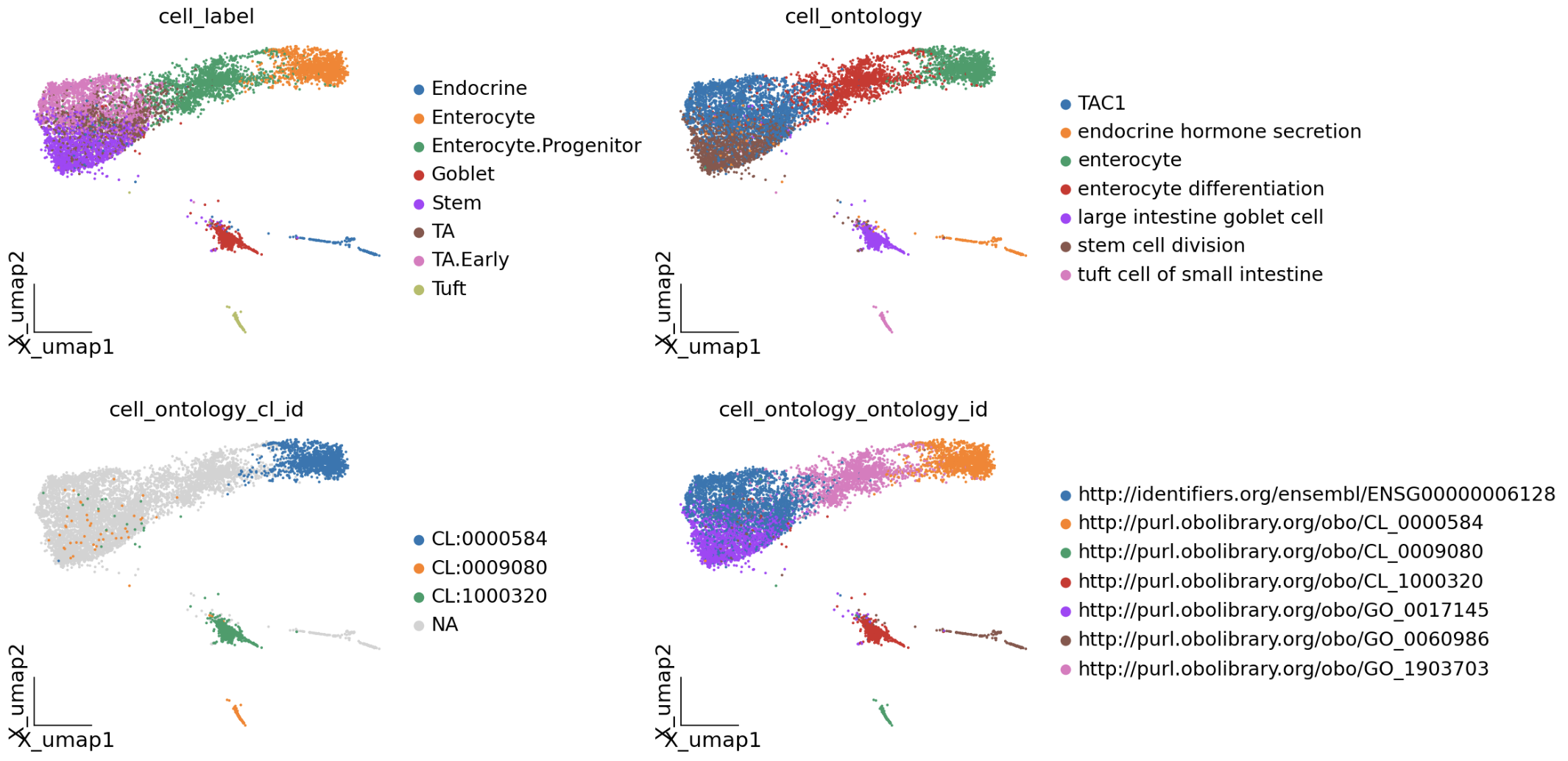
🤖 使用LLM辅助细胞类型映射
在实际工作中,研究者经常使用缩写来命名细胞类型(比如TA代表Transit Amplifying cell,EC代表Endothelial cell)。这些缩写可能会影响与细胞本体论的匹配效果。为解决这个问题,我们创新性地引入了LLM(大语言模型)来智能解析这些缩写。
配置参数说明:
| 参数 | 说明 | 示例 |
|---|---|---|
| api_type | API类型 | openai, anthropic, ollama |
| tissue_context | 组织来源 | "gut", "brain", "liver" |
| species | 研究物种 | "mouse", "human", "rat" |
| study_context | 研究背景 | "肠道上皮细胞单细胞测序" |
| api_key | API密钥 | "sk-..." |
⚠️ 安全提示:请妥善保管你的API密钥,不要将其暴露在公开环境中。
mapper.setup_llm_expansion(
api_type="openai", model='gpt-4o-2024-11-20',
tissue_context="gut", # 组织上下文
species="mouse", # 物种信息
study_context="Epithelial cells from the small intestine and organoids of mice. Some of the cells were also subject to Salmonella or Heligmosomoides polygyrus infection",
api_key="sk-*"
)
✓ Loaded 10 cached abbreviation expansions
✓ LLM expansion functionality setup complete (Type: openai, Model: gpt-4o-2024-11-20)
🧬 Tissue context: gut
🔬 Study context: Epithelial cells from the small intestine and organoids of mice. Some of the cells were also subject to Salmonella or Heligmosomoides polygyrus infection
🐭 Species: mouse
你可以选择任何符合openai规则的api作为输入,例如ohmygpt.
mapper.setup_llm_expansion(
api_type="custom_openai",
api_key="sk-*",
model="gpt-4.1-2025-04-14",
base_url="https://api.ohmygpt.com/v1"
)
大语言模型辅助映射
mapping_results = mapper.map_adata_with_expansion(
adata=adata,
cell_name_col='cell_label',
threshold=0.5,
expand_abbreviations=True # 启用缩写扩展
)
mapper.print_mapping_summary(mapping_results, top_n=15)
🔤 Identified potential abbreviation: Stem
🔤 Identified potential abbreviation: TA.Early
🔤 Identified potential abbreviation: TA
🔤 Identified potential abbreviation: Tuft
🔤 Identified potential abbreviation: Goblet
🤖 Expanding 5 abbreviations using LLM...
📝 [1/5] Expanding: Stem
✓ → Intestinal stem cell (Confidence: high)
💡 Alternatives: Stem cell, Crypt stem cell
📝 [2/5] Expanding: TA.Early
✓ → Transit Amplifying Early cell (Confidence: high)
💡 Alternatives: Transit Amplifying progenitor cell (early stage), Transient Amplifying Early cell
📝 [3/5] Expanding: TA
✓ → Transit amplifying cell (Confidence: high)
💡 Alternatives: Tumor-associated cell, T cell activation-related cell
📝 [4/5] Expanding: Tuft
✓ → Tuft cell (Confidence: high)
💡 Alternatives: Brush cell
📝 [5/5] Expanding: Goblet
✓ → Goblet cell (Confidence: high)
✓ Tuft -> tuft cell (Similarity: 0.787)
✓ Enterocyte -> enterocyte (Similarity: 0.776)
✓ TA -> transit amplifying cell of appendix (Similarity: 0.741)
✓ Stem -> intestinal crypt stem cell (Similarity: 0.735)
✓ Goblet -> small intestine goblet cell (Similarity: 0.734)
✓ Enterocyte.Progenitor -> enterocyte differentiation (Similarity: 0.688)
✓ TA.Early -> transit amplifying cell (Similarity: 0.688)
✓ Endocrine -> endocrine hormone secretion (Similarity: 0.643)
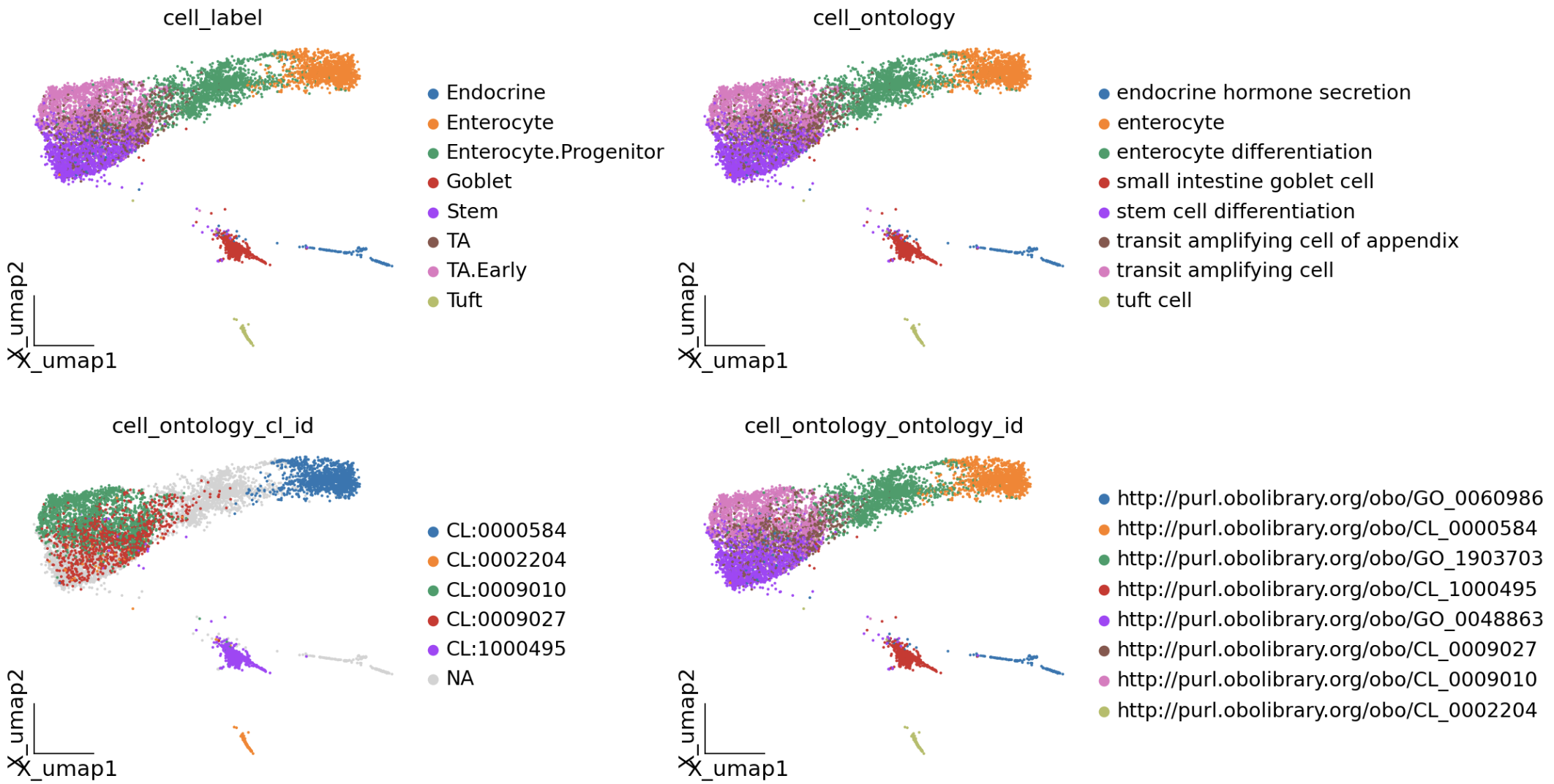
我们可以发现TA 和 TA.Early在扩写了细胞名称后,被成功映射到了对应的细胞。
adata.obs[['cell_label','cell_ontology','cell_ontology_similarity',
'cell_ontology_ontology_id','cell_ontology_ontology_id','cell_ontology_cl_id']].head()
| cell_label | cell_ontology | cell_ontology_similarity | cell_ontology_ontology_id | cell_ontology_ontology_id | cell_ontology_cl_id | |
|---|---|---|---|---|---|---|
| index | ||||||
| B1_AAACATACCACAAC_Control_Enterocyte.Progenitor | Enterocyte.Progenitor | enterocyte differentiation | 0.688446 | http://purl.obolibrary.org/obo/GO_1903703 | http://purl.obolibrary.org/obo/GO_1903703 | None |
| B1_AAACGCACGAGGAC_Control_Stem | Stem | intestinal crypt stem cell | 0.735365 | http://purl.obolibrary.org/obo/CL_0002250 | http://purl.obolibrary.org/obo/CL_0002250 | CL:0002250 |
| B1_AAACGCACTAGCCA_Control_Stem | Stem | intestinal crypt stem cell | 0.735365 | http://purl.obolibrary.org/obo/CL_0002250 | http://purl.obolibrary.org/obo/CL_0002250 | CL:0002250 |
| B1_AAACGCACTGTCCC_Control_Stem | Stem | intestinal crypt stem cell | 0.735365 | http://purl.obolibrary.org/obo/CL_0002250 | http://purl.obolibrary.org/obo/CL_0002250 | CL:0002250 |
| B1_AAACTTGACCACCT_Control_Enterocyte.Progenitor | Enterocyte.Progenitor | enterocyte differentiation | 0.688446 | http://purl.obolibrary.org/obo/GO_1903703 | http://purl.obolibrary.org/obo/GO_1903703 | None |
🧬 细胞分类学注释
我们在前面的分析,已经很好地获得了一系列合理的标准的细胞名,不过,我们还可以使用另外一个数据库,来获取不同的严格细胞名。
我们可以手动从国家生物信息学中心数据库进行数据下载:https://download.cncb.ac.cn/celltaxonomy/Cell_Taxonomy_resource.txt
!wget https://download.cncb.ac.cn/celltaxonomy/Cell_Taxonomy_resource.txt -O new_ontology/Cell_Taxonomy_resource.txt
然后,我们需要加载我们的数据库
mapper.load_cell_taxonomy_resource("new_ontology/Cell_Taxonomy_resource.txt",
species_filter=["Homo sapiens", "Mus musculus"])
📊 Loading Cell Taxonomy resource from: new_ontology/Cell_Taxonomy_resource.txt
✓ Loaded 226222 taxonomy entries
🐭 Filtered by species ['Homo sapiens', 'Mus musculus']: 224736/226222 entries
🧠 Creating embeddings for 2540 taxonomy cell types...
Batches: 100%
80/80 [00:00<00:00, 307.18it/s]
✓ Created taxonomy embeddings for 2540 cell types
📈 Species distribution:
🐭 Mus musculus: 141727 entries
🐭 Homo sapiens: 83009 entries
🧬 Unique cell types: 2540
🎯 Unique markers: 25818
与前面的分析类似,我们这里也提供了一个一键mapping函数:
enhanced_results = mapper.map_adata_with_taxonomy(
adata,
cell_name_col="cell_label",
new_col_name="enhanced_cell_ontology",
expand_abbreviations=True,
use_taxonomy=True,
species="Mus musculus",
tissue_context="Gut",
threshold=0.3,
)
mapper.print_mapping_summary_taxonomy(enhanced_results)

结果非常漂亮:
================================================================================
ENHANCED MAPPING SUMMARY (ONTOLOGY + TAXONOMY)
================================================================================
Total mappings: 8
High confidence: 8 (100.00%)
Low confidence: 0
Average similarity: 0.724
LLM expansions: 5
Taxonomy enhanced: 8
DETAILED MAPPING RESULTS (Top 10)
--------------------------------------------------------------------------------
1. [✓] Tuft
🔤 Expanded: Tuft → Tuft cell
🎯 Ontology: tuft cell
Similarity: 0.787
CL ID: CL:0002204
🧬 Taxonomy: Intestinal tuft cell
Similarity: 0.814
Matched from: Tuft cell
CT ID: CT:00002708
🎯 Marker: Dclk1
🧬 Gene: 1700113D08Rik,2810480F11Rik,AI836758,CPG1,Clic,Click-I,Cpg16,Dc,Dcamk,Dcamkl1,Dcl,Dclk,mKIAA0369,DCLK1
🆔 ENTREZ: 13175
2. [✓] Enterocyte
🎯 Ontology: enterocyte
Similarity: 0.776
CL ID: CL:0000584
🧬 Taxonomy: Enterocyte
Similarity: 1.000
Matched from: Enterocyte
CT ID: CT:00000594
🎯 Marker: Btnl1
🧬 Gene: Btn,Btnl3,Gm316,Gm33,NG1
🆔 ENTREZ: 100038862
3. [✓] TA
🔤 Expanded: TA → Transit amplifying cell
🎯 Ontology: transit amplifying cell of appendix
Similarity: 0.741
CL ID: CL:0009027
🧬 Taxonomy: Transit amplifying cell
Similarity: 1.000
Matched from: Transit amplifying cell
CT ID: CT:00001800
🎯 Marker: Rpl18a
🧬 Gene: 2510019J09Rik
🆔 ENTREZ: 76808
4. [✓] Stem
🔤 Expanded: Stem → Intestinal stem cell
🎯 Ontology: intestinal crypt stem cell
Similarity: 0.735
CL ID: CL:0002250
🧬 Taxonomy: Intestinal stem cell
Similarity: 1.000
Matched from: Intestinal stem cell
CT ID: CT:00000029
🎯 Marker: Alcam
🧬 Gene: AI853494,BE,BEN,CD166,DM-G,DM-GRASP,MuS,MuSC,SC,SC1,ALCAM
🆔 ENTREZ: 11658
5. [✓] Goblet
🔤 Expanded: Goblet → Goblet cell
🎯 Ontology: small intestine goblet cell
Similarity: 0.734
CL ID: CL:1000495
🧬 Taxonomy: Goblet cell
Similarity: 1.000
Matched from: Goblet cell
CT ID: CT:00000223
🎯 Marker: Gal3st2b
🧬 Gene: Gal3ST-2,Gal3st2,Gm9994
🆔 ENTREZ: 100041596
6. [✓] Enterocyte.Progenitor
🎯 Ontology: enterocyte differentiation
Similarity: 0.688
CL ID: None
🧬 Taxonomy: Enterocyte progenitor cell
Similarity: 0.898
Matched from: Enterocyte.Progenitor
CT ID: CT:00001880
🎯 Marker: CD24
🧬 Gene: CD24A
🆔 ENTREZ: 100133941
7. [✓] TA.Early
🔤 Expanded: TA.Early → Transit Amplifying Early Cell
🎯 Ontology: transit amplifying cell
Similarity: 0.688
CL ID: CL:0009010
🧬 Taxonomy: Transit amplifying cell
Similarity: 0.894
Matched from: Transit Amplifying Early Cell
CT ID: CT:00001800
🎯 Marker: Rpl18a
🧬 Gene: 2510019J09Rik
🆔 ENTREZ: 76808
8. [✓] Endocrine
🎯 Ontology: endocrine hormone secretion
Similarity: 0.643
CL ID: None
🧬 Taxonomy: Endocrine cell
Similarity: 0.816
Matched from: Endocrine
CT ID: CT:00000227
🎯 Marker: Cyb5r3
🧬 Gene: 0610016L08Rik,2500002N19Rik,B5R,C85115,Di,Dia,Dia-1,Dia1,WU:Cyb5r3
🆔 ENTREZ: 109754
================================================================================
🔍 映射结果检验
为确保映射结果的准确性,我们提供了多种验证方法:
- 手动查询匹配结果
- 检查相似度分数
- 验证本体论ID
实用技巧:
- 相似度分数 > 0.7:高度可信
- 相似度分数 0.5-0.7:需要人工核验
- 相似度分数 < 0.5:建议使用LLM辅助或手动映射
res=mapper.find_similar_cells("T helper cell", top_k=10)
🎯 Ontology cell types most similar to 'T helper cell':
1. helper T cell (Similarity: 0.780)
2. T-helper 1 cell activation (Similarity: 0.738)
3. T-helper 2 cell activation (Similarity: 0.709)
4. T-helper 9 cell (Similarity: 0.707)
5. T-helper 2 cell (Similarity: 0.690)
6. T-helper 1 cell (Similarity: 0.687)
7. T cell domain (Similarity: 0.678)
8. regulation of T-helper 1 cell activation (Similarity: 0.675)
9. CD4-positive helper T cell (Similarity: 0.664)
10. T-helper 1 cell cytokine production (Similarity: 0.660)
res=mapper.find_similar_cells_taxonomy("T helper cell", top_k=2)
🧬 Taxonomy cell types most similar to 'T helper cell':
1. Helper T cell (Similarity: 0.966)
🐭 Species: Mus musculus
🎯 Marker: Tigit
🆔 CT ID: CT:00000919
2. T-helper 1 cell (Similarity: 0.926)
🐭 Species: Homo sapiens
🎯 Marker: CXCR6
🆔 CT ID: CT:00000502
获取本体论中的细胞信息
mapper.get_cell_info("regulatory T cell")
ℹ️ === regulatory T cell ===
🆔 Ontology ID: http://purl.obolibrary.org/obo/CL_0000815
📝 Description: regulatory T cell: A T cell which regulates overall immune responses as well as the responses of other T cell subsets through direct cell-cell contact and cytokine release. This cell type may express FoxP3 and CD25 and secretes IL-10 and TGF-beta.
获取本体论中的细胞信息
mapper.get_cell_info("regulatory T cells")
✗ Cell type not found: regulatory T cells
🔍 Found 0 cell types containing 'regulatory t cells':
获取本体论中的细胞类别的信息
my_categories = ["immune cell", "epithelial"]
mapper.browse_ontology_by_category(categories=my_categories, max_per_category=5)
📂 === Browse Ontology Cell Types by Category ===
🔍 Found 0 cell types containing 'immune cell':
--------------------------------------------------
🔍 Found 395 cell types containing 'epithelial':
1. NS forest marker set of airway submucosal gland collecting duct epithelial cell (Human Lung).
2. epithelial fate stem cell
3. epithelial cell
4. ciliated epithelial cell
5. duct epithelial cell
... 390 more results
🏷️ 【epithelial related】 (Showing top 5):
1. NS forest marker set of airway submucosal gland collecting duct epithelial cell (Human Lung).
2. epithelial fate stem cell
3. epithelial cell
4. ciliated epithelial cell
5. duct epithelial cell
--------------------------------------------------
查看本体论中的细胞信息
# 查看前50个细胞类型
res=mapper.list_ontology_cells(max_display=10)
📊 Total 16841 cell types in ontology
📋 First 10 cell types:
1. TAC1
2. STAB1
3. TLL1
4. MSR1
5. TNC
6. ROS1
7. TNIP3
8. HOMER3
9. FCGR2B
10. BPIFB2
... 16831 more cell types
💡 Use return_all=True to get complete list
了解本体论的整体情况
# 了解本体论的整体情况
stats = mapper.get_ontology_statistics()
📊 === Ontology Statistics ===
📝 Total cell types: 16841
📏 Average name length: 31.7 characters
📏 Shortest name length: 3 characters
📏 Longest name length: 144 characters
🔤 Most common words:
of: 5473 times
cell: 3857 times
regulation: 3168 times
negative: 1009 times
positive: 1003 times
process: 980 times
development: 875 times
differentiation: 727 times
muscle: 639 times
in: 571 times
📈 总结与展望
通过使用细胞本体论进行细胞类型映射,我们可以:
- 🎯 实现细胞类型注释的标准化
- 📊 提高数据的可比性和可重复性
- 🔗 促进不同数据集之间的整合分析
- 🧬 发现细胞类型之间的生物学联系
最佳实践建议:
- 在发表论文时,同时提供原始注释和映射后的细胞本体论ID
- 定期更新细胞本体论数据库,保持与最新研究同步
- 建立标准化的细胞类型注释流程,提高研究效率
🎉 恭喜你完成了本教程!现在你已经掌握了专业的细胞类型映射方法。
本文来自博客园,作者:Starlitnightly,转载请注明原文链接:https://www.cnblogs.com/starlitnightly/p/18919221



 浙公网安备 33010602011771号
浙公网安备 33010602011771号