JCVI作图进阶系列(四)
Let's plot the arabidopsis genome.
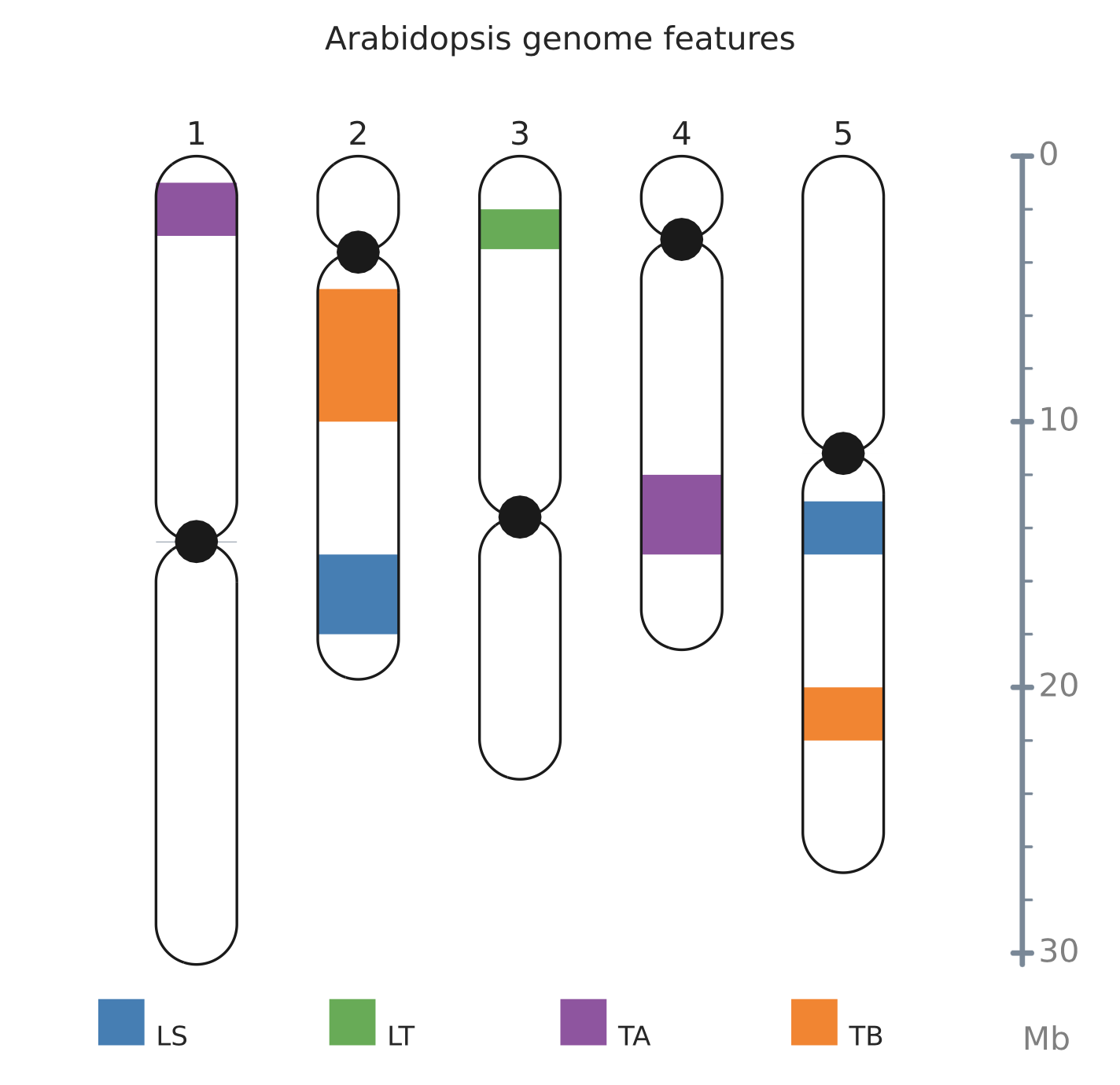
可用于集中展示着丝粒以及端粒区域重复单元丰度。
一、数据准备
#make chromosome size
vim athaliana.sizes
*********************
Chr1 30427671
Chr2 19698289
Chr3 23459830
Chr4 18585056
Chr5 26975502
*********************#add a number of interesting features
vim features.bed
*********************
Chr1 1000000 3000000 type A
Chr4 12000000 15000000 type A
Chr2 5000000 10000000 type B
Chr5 20000000 22000000 type B
Chr2 15000000 18000000 LINE/SINE
Chr5 13000000 15000000 LINE/SINE
Chr3 2000000 3500000 LTR
Chr1 14511721 14538721 centromere
Chr2 3611838 3611883 centromere
Chr3 13589756 13589816 centromere
Chr4 3133663 3133674 centromere
Chr5 11194537 11194848 centromere
*********************#change labels and colors
vim athaliana.idmap
*********************
LINE/SINE LS #377EB8
LTR LT #4DAF4A
type A TA #984EA3
type B TB #FF7F00
*********************二、一键绘图
python -m jcvi.graphics.chromosome features.bed athaliana.idmap \
--size=athaliana.sizes --title="Arabidopsis genome features" --gauge --notex三、结果展示
字体等其他设置可以导入AI再优化调整




 浙公网安备 33010602011771号
浙公网安备 33010602011771号