plink 格式中提取包含杂合位点的SNP位点
001、
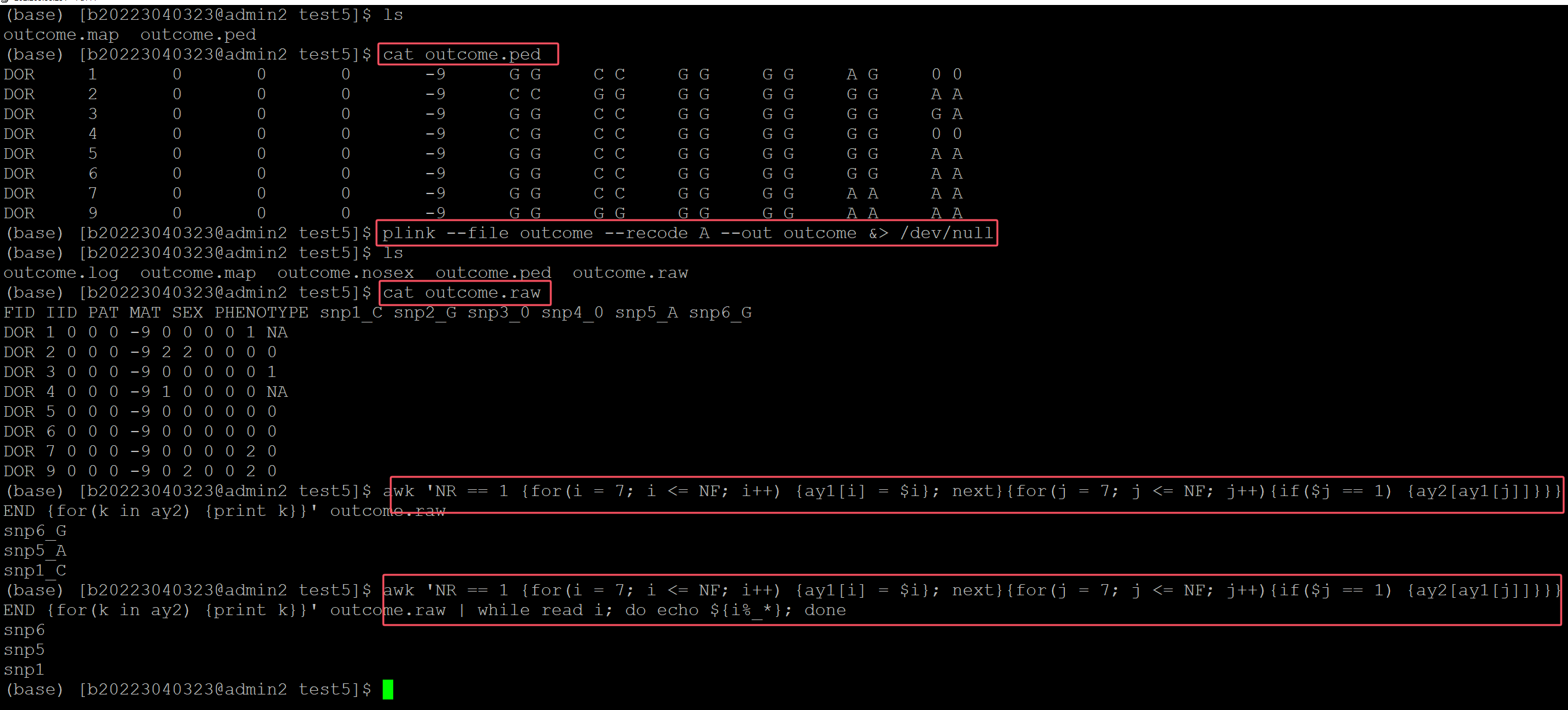
(base) [b20223040323@admin2 test5]$ ls ## 测试数据 outcome.map outcome.ped (base) [b20223040323@admin2 test5]$ cat outcome.ped DOR 1 0 0 0 -9 G G C C G G G G A G 0 0 DOR 2 0 0 0 -9 C C G G G G G G G G A A DOR 3 0 0 0 -9 G G C C G G G G G G G A DOR 4 0 0 0 -9 C G C C G G G G G G 0 0 DOR 5 0 0 0 -9 G G C C G G G G G G A A DOR 6 0 0 0 -9 G G C C G G G G G G A A DOR 7 0 0 0 -9 G G C C G G G G A A A A DOR 9 0 0 0 -9 G G G G G G G G A A A A (base) [b20223040323@admin2 test5]$ plink --file outcome --recode A --out outcome &> /dev/null ## 转换格式,minor allele 为0, major alles 为2, het为1 (base) [b20223040323@admin2 test5]$ ls outcome.log outcome.map outcome.nosex outcome.ped outcome.raw (base) [b20223040323@admin2 test5]$ cat outcome.raw FID IID PAT MAT SEX PHENOTYPE snp1_C snp2_G snp3_0 snp4_0 snp5_A snp6_G DOR 1 0 0 0 -9 0 0 0 0 1 NA DOR 2 0 0 0 -9 2 2 0 0 0 0 DOR 3 0 0 0 -9 0 0 0 0 0 1 DOR 4 0 0 0 -9 1 0 0 0 0 NA DOR 5 0 0 0 -9 0 0 0 0 0 0 DOR 6 0 0 0 -9 0 0 0 0 0 0 DOR 7 0 0 0 -9 0 0 0 0 2 0 DOR 9 0 0 0 -9 0 2 0 0 2 0 ## 利用awk命令进行统计 (base) [b20223040323@admin2 test5]$ awk 'NR == 1 {for(i = 7; i <= NF; i++) {ay1[i] = $i}; next}{for(j = 7; j <= NF; j++){if($j == 1) {ay2[ay1[j]]}}} END {for(k in ay2) {print k}}' outcome.raw snp6_G snp5_A snp1_C (base) [b20223040323@admin2 test5]$ awk 'NR == 1 {for(i = 7; i <= NF; i++) {ay1[i] = $i}; next}{for(j = 7; j <= NF; j++){if($j == 1) {ay2[ay1[j]]}}} END {for(k in ay2) {print k}}' outcome.raw | while read i; do echo ${i%_*}; done snp6 snp5 snp1
。




 浙公网安备 33010602011771号
浙公网安备 33010602011771号