R语言中绘制火山图
001、
# 生成随机数据 set.seed(123) # 设置种子以便结果可重复 genes <- paste("Gene", 1:1000) # 基因名称 logFC <- rnorm(1000, mean = 0, sd = 2) # log2 fold change,均值为0,标准差为2的正态分布 pvalue <- runif(1000, min = 0, max = 1) # p值,0到1之间的均匀分布 genes logFC pvalue # 创建数据框 data <- data.frame(genes = genes, logFC = logFC, pvalue = pvalue) head(data) # 添加一列来标记显著性 data$significant <- ifelse(abs(data$logFC) > 1 & data$pvalue < 0.05, "Significant", "Not Significant") head(data) # 绘制火山图 ggplot(data, aes(x = logFC, y = -log10(pvalue), color = significant)) + geom_point(alpha = 0.6, size = 1.5) + # 绘制散点,alpha控制透明度,size控制点的大小 scale_color_manual(values = c("grey", "red")) + # 自定义颜色,显著基因用红色 theme_minimal() + # 使用简单的主题风格 labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + # 添加标题和轴标签 theme(plot.title = element_text(hjust = 0.5)) # 将标题居中 ggplot(data, aes(x = logFC, y = -log10(pvalue), color = significant)) + geom_point(alpha = 0.6, size = 1.5) + scale_color_manual(values = c("grey", "red")) + theme_minimal() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + theme(plot.title = element_text(hjust = 0.5)) + geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "blue") + # 添加竖线,表示logFC的阈值 geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "blue") # 添加横线,表示p-value的阈值 # 根据logFC和p-value进一步分类 data$expression <- ifelse(data$logFC > 1 & data$pvalue < 0.05, "Upregulated", ifelse(data$logFC < -1 & data$pvalue < 0.05, "Downregulated", "Not Significant")) # 查看数据框的前几行,确认分类是否正确 head(data) ggplot(data, aes(x = logFC, y = -log10(pvalue), color = expression)) + geom_point(alpha = 0.6, size = 1.5) + # 绘制散点,透明度和点的大小 scale_color_manual(values = c("blue", "grey", "red")) + # 自定义颜色:下调为蓝色,不显著为灰色,上调为红色 theme_minimal() + # 简洁主题 labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + # 添加标题和轴标签 theme(plot.title = element_text(hjust = 0.5)) + # 标题居中 geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "black") + # 添加阈值竖线 geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") # 添加阈值横线 # 安装 ggrepel 包 # install.packages("ggrepel") # 加载 ggrepel 包 library(ggrepel) # 只标记显著的上调和下调基因 significant_genes <- data[data$expression != "Not Significant", ] # 绘制带基因标记的火山图 ggplot(data, aes(x = logFC, y = -log10(pvalue), color = expression)) + geom_point(alpha = 0.6, size = 1.5) + scale_color_manual(values = c("blue", "grey", "red")) + theme_minimal() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + theme(plot.title = element_text(hjust = 0.5)) + geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "black") + geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") + #避免标签重叠的函数,可以让基因名称显得更加清晰。 geom_text_repel(data = significant_genes, aes(label = genes), size = 3, box.padding = 0.3, point.padding = 0.3) ggplot(data, aes(x = logFC, y = -log10(pvalue), color = expression)) + geom_point(aes(size = -log10(pvalue), alpha = abs(logFC))) + # 点的大小和透明度。 #size = -log10(pvalue):显著性越高的点越大。 #alpha = abs(logFC):log2 fold change 越大,点越不透明。 scale_color_manual(values = c("blue", "grey", "red")) + theme_minimal() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + theme(plot.title = element_text(hjust = 0.5)) + geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "black") + geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") # 为数据添加一个模拟条件列 data$condition <- sample(c("Condition A", "Condition B"), nrow(data), replace = TRUE) ggplot(data, aes(x = logFC, y = -log10(pvalue), color = expression)) + geom_point(alpha = 0.6, size = 1.5) + scale_color_manual(values = c("blue", "grey", "red")) + theme_minimal() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + theme(plot.title = element_text(hjust = 0.5)) + geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "black") + geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") + facet_wrap(~ condition) # 按条件分面显示 # 安装 plotly 包 # install.packages("plotly") # 加载 plotly 包 library(plotly) p <- ggplot(data, aes(x = logFC, y = -log10(pvalue), color = expression)) + geom_point(alpha = 0.6, size = 1.5) + scale_color_manual(values = c("blue", "grey", "red")) + theme_minimal() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 P-value") + theme(plot.title = element_text(hjust = 0.5)) + geom_vline(xintercept = c(-1, 1), linetype = "dashed", color = "black") + geom_hline(yintercept = -log10(0.05), linetype = "dashed", color = "black") # 转换为交互式图形 ggplotly(p)
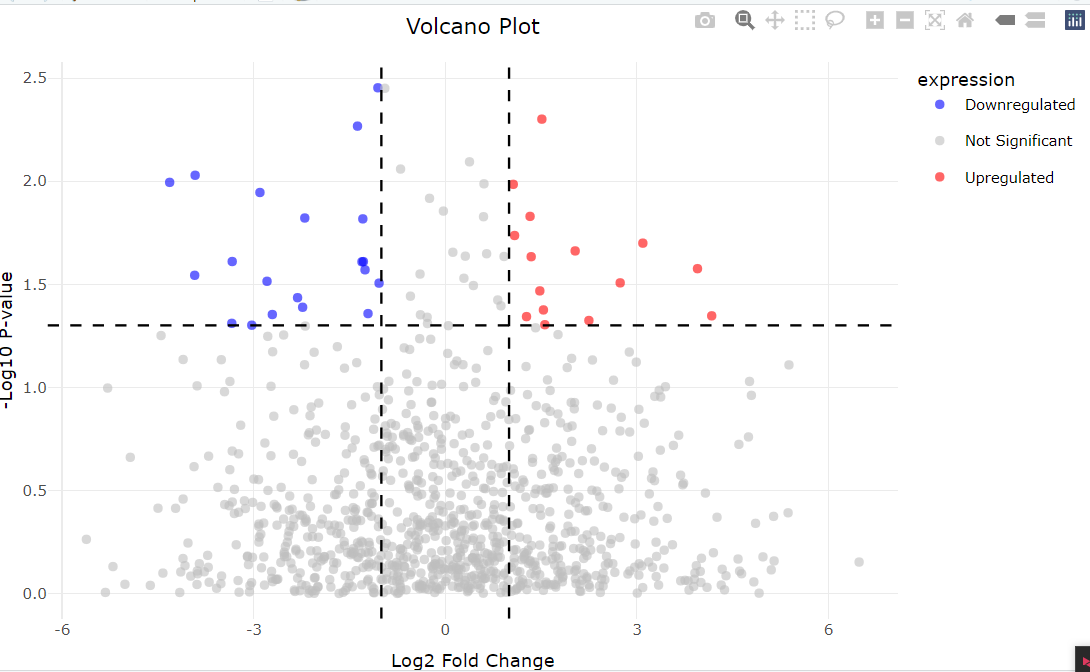
。
source:
01、https://blog.csdn.net/zegeai/article/details/142934945




 浙公网安备 33010602011771号
浙公网安备 33010602011771号