使用PopLDdecay进行连锁不平衡分析
001、 软件下载:
github网站:https://github.com/BGI-shenzhen/PopLDdecay
002、下载最新版本
wget -c https://github.com/BGI-shenzhen/PopLDdecay/archive/refs/tags/v3.43.tar.gz
003、解压、编译安装
tar -xzvf PopLDdecay-3.43.tar.gz
cd PopLDdecay-3.43/
chmod +x configure
./configure
make
mv PopLDdecay bin/
cd test/
./PopLDdecay
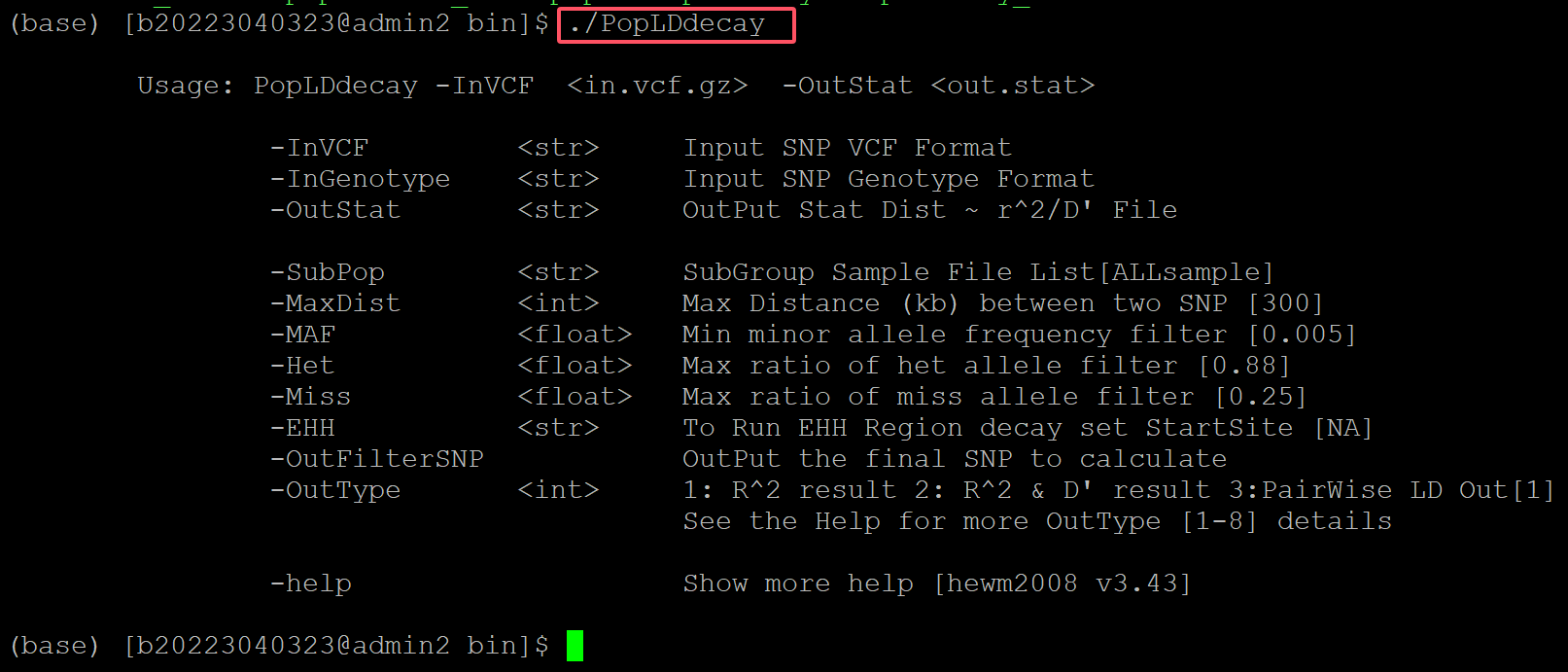
.
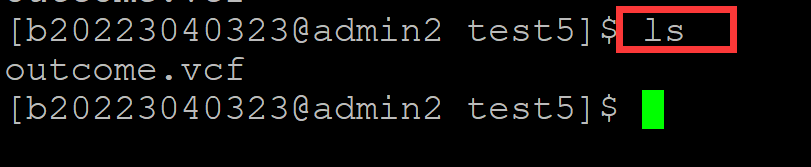
004、准备测试文件
[b20223040323@admin2 test5]$ ls
outcome.vcf
005、单群体运算
PopLDdecay -InVCF ./outcome.vcf -MaxDist 1000 -OutType 3 -OutStat out
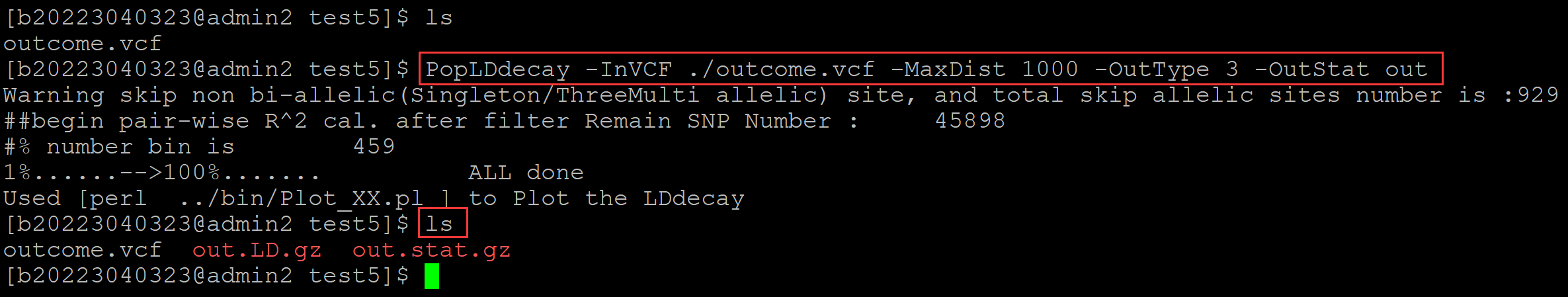
。
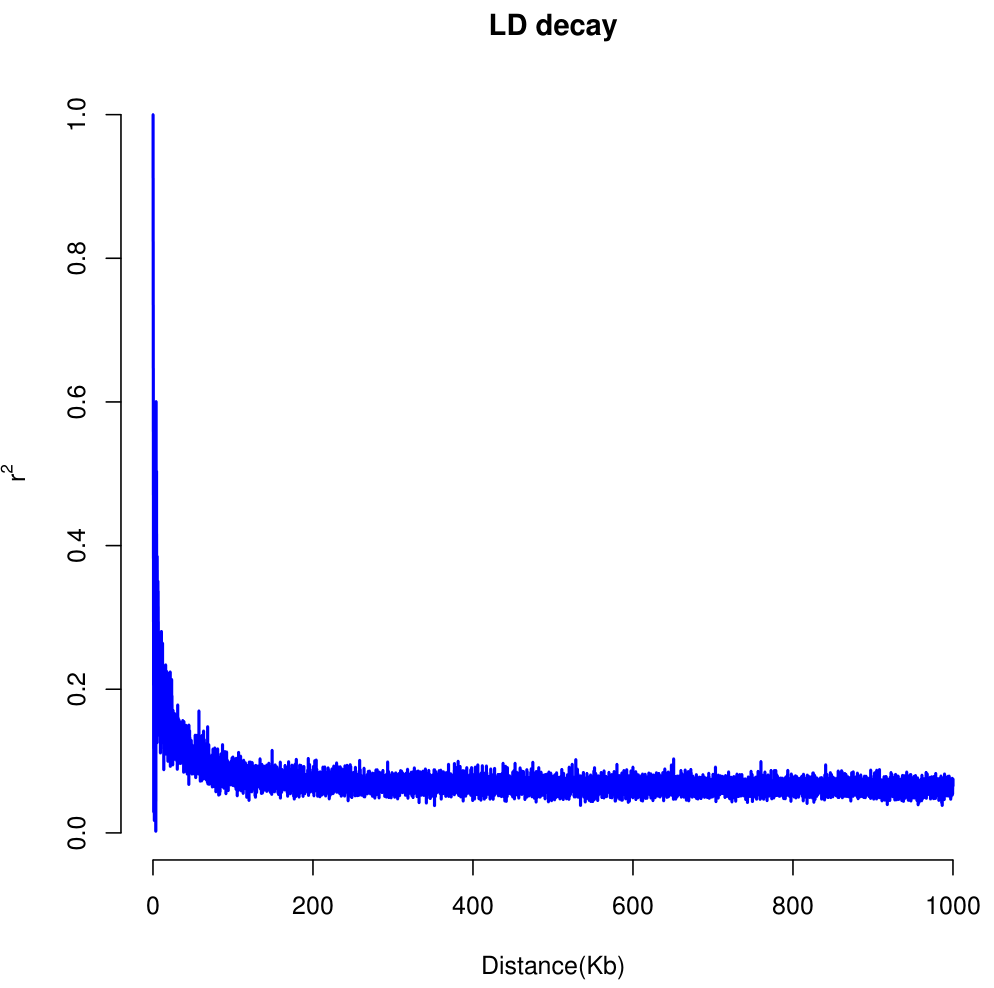
006、可视化
[b20223040323@admin2 test5]$ ls outcome.vcf out.LD.gz out.stat.gz snp.bin snp.r [b20223040323@admin2 test5]$ Plot_OnePop.pl -inFile out.stat.gz -bin1 10 -bin2 100 -break 100 -output snp null device 1 null device 1
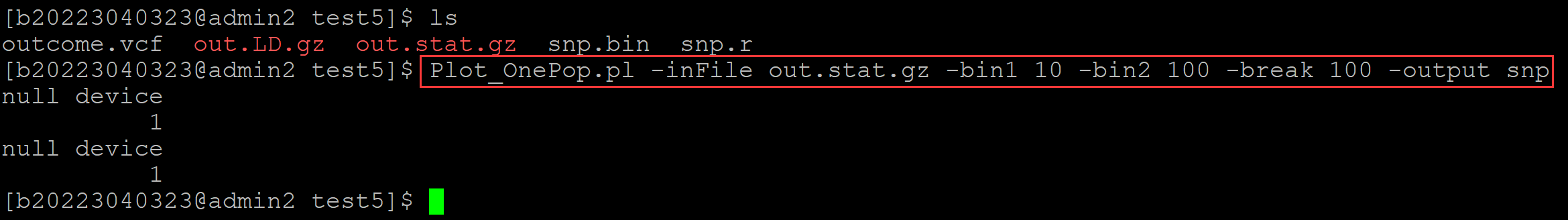
。
007、
008、多群体的计算
[b20223040323@admin2 test5]$ ls dor.list gmm.list outcome.vcf sun.list [b20223040323@admin2 test5]$ head -n 3 dor.list DOR1 DOR2 DOR3 [b20223040323@admin2 test5]$ PopLDdecay -InVCF outcome.vcf -OutStat dor.stat.gz -SubPop dor.list the Number of subPop samples[found in VCF] is 10 Warning skip non bi-allelic(Singleton/ThreeMulti allelic) site, and total skip allelic sites number is :7403 ##begin pair-wise R^2 cal. after filter Remain SNP Number : 39246 #% number bin is 393 1%......-->100%....... ALL done Used [perl ../bin/Plot_XX.pl ] to Plot the LDdecay [b20223040323@admin2 test5]$ PopLDdecay -InVCF outcome.vcf -OutStat gmm.stat.gz -SubPop gmm.list the Number of subPop samples[found in VCF] is 10 Warning skip non bi-allelic(Singleton/ThreeMulti allelic) site, and total skip allelic sites number is :5552 ##begin pair-wise R^2 cal. after filter Remain SNP Number : 41071 #% number bin is 411 1%......-->100%....... ALL done Used [perl ../bin/Plot_XX.pl ] to Plot the LDdecay [b20223040323@admin2 test5]$ PopLDdecay -InVCF outcome.vcf -OutStat sun.stat.gz -SubPop sun.list the Number of subPop samples[found in VCF] is 10 Warning skip non bi-allelic(Singleton/ThreeMulti allelic) site, and total skip allelic sites number is :3423 ##begin pair-wise R^2 cal. after filter Remain SNP Number : 43219 #% number bin is 433 1%......-->100%....... ALL done Used [perl ../bin/Plot_XX.pl ] to Plot the LDdecay [b20223040323@admin2 test5]$ ls dor.list dor.stat.gz gmm.list gmm.stat.gz outcome.vcf sun.list sun.stat.gz
。
009、可视化
[b20223040323@admin2 test5]$ ls dor.list dor.stat.gz gmm.list gmm.stat.gz multi.list outcome.vcf sun.list sun.stat.gz [b20223040323@admin2 test5]$ cat multi.list ## 准备一个配置文件 dor.stat.gz dor gmm.stat.gz gmm sun.stat.gz sun [b20223040323@admin2 test5]$ Plot_MultiPop.pl -inList multi.list --out result_pig -bin1 10 -bin2 500 ## 绘图 null device 1 null device 1 [b20223040323@admin2 test5]$ ls dor.list gmm.list multi.list result_pig.dor result_pig.pdf result_pig.sun sun.stat.gz dor.stat.gz gmm.stat.gz outcome.vcf result_pig.gmm result_pig.png sun.list
.
备用参数:
01、Plot_MultiPop.pl -inList multi.list --out result_pig -bin1 10 -bin2 100 -break 100
02、Plot_MultiPop.pl -inList multi.list --out result_pig -bin1 20 -bin2 50 -break 1000
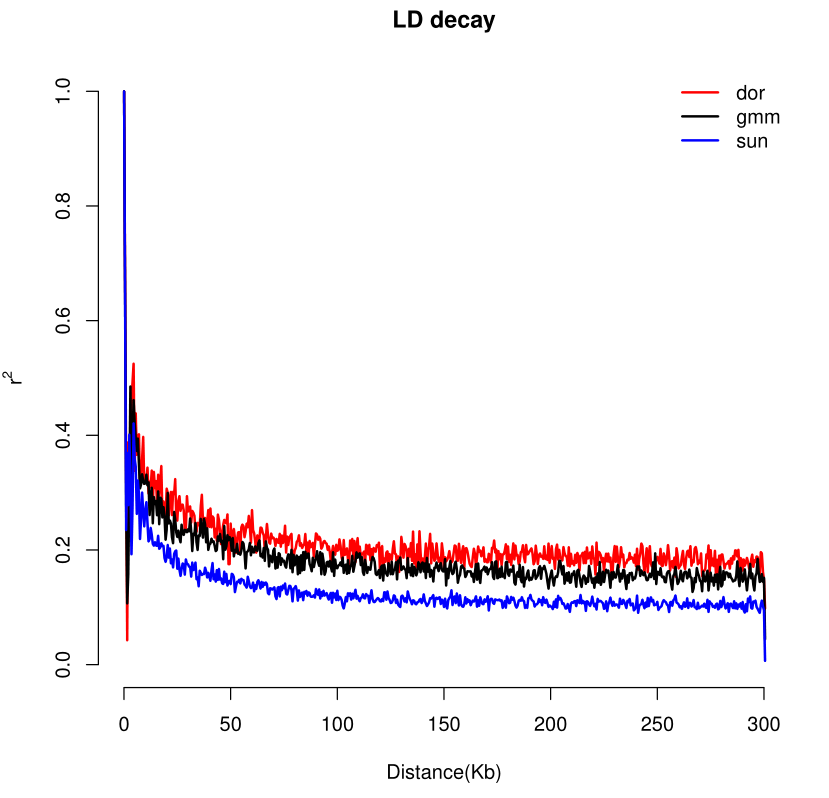
绘图结果:
。
ref:
01、https://cloud.tencent.com/developer/article/1677015




 浙公网安备 33010602011771号
浙公网安备 33010602011771号