R语言中统计基因型数据中每行中各种碱基的数目
001、
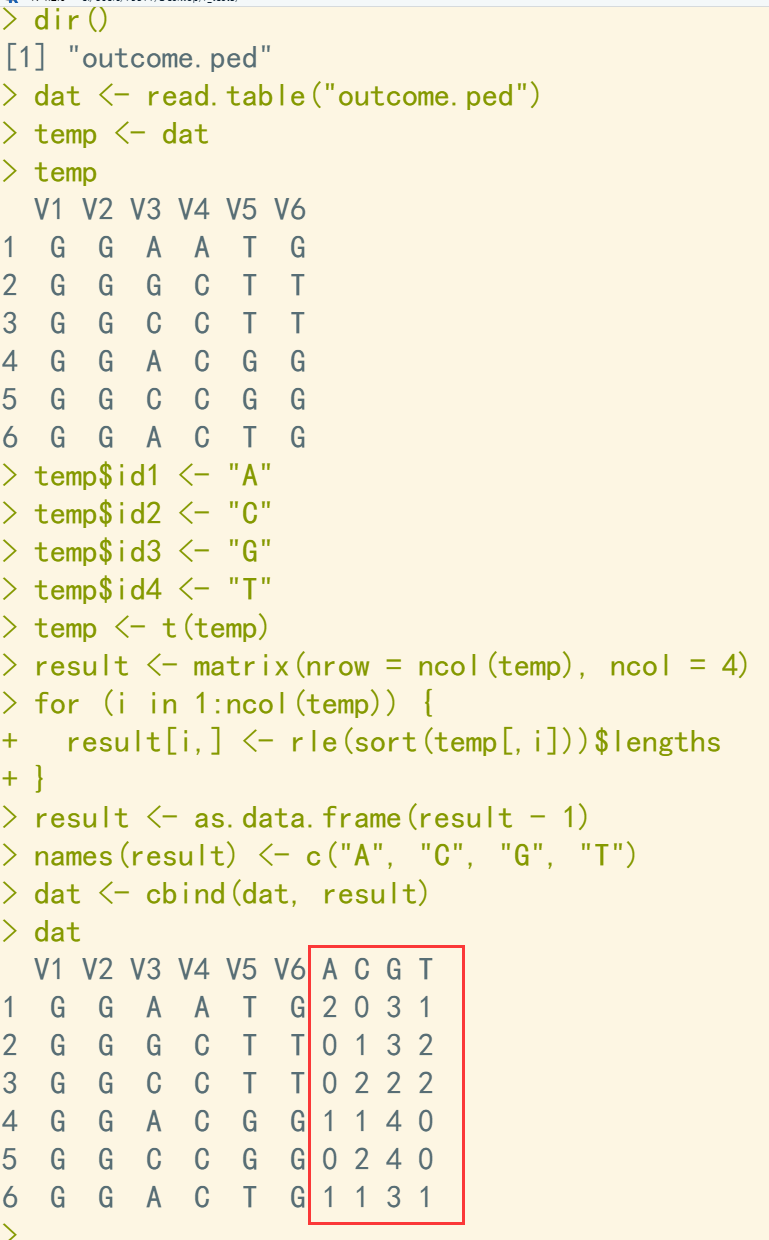
dir() dat <- read.table("outcome.ped") temp <- dat temp temp$id1 <- "A" temp$id2 <- "C" temp$id3 <- "G" temp$id4 <- "T" temp <- t(temp) result <- matrix(nrow = ncol(temp), ncol = 4) for (i in 1:ncol(temp)) { result[i,] <- rle(sort(temp[,i]))$lengths ## 统计每列中各种字符的次数 } result <- as.data.frame(result - 1) names(result) <- c("A", "C", "G", "T") dat <- cbind(dat, result) ## 跟原始数据合并 dat
002、pyhton实现
root@PC1:/home/test3# ls outcome.ped test.py root@PC1:/home/test3# cat outcome.ped G G A A T G G G G C T T G G C C T T G G A C G G G G C C G G G G A C T G root@PC1:/home/test3# cat test.py ## 实现脚本 #!/usr/bin/python in_file = open("outcome.ped", "r") out_file = open("result.txt", "w") lines = in_file.readlines() for i in lines: temp = i.split() A = 0; C = 0; G = 0; T = 0; temp_list = [] for j in range(len(temp)): if temp[j] == "A": A = A + 1 else: if temp[j] == "C": C = C + 1 else: if temp[j] == "G": G = G + 1 else: T = T + 1 temp_list.extend([str(A),str(C),str(G),str(T)]) out_file.write(' '.join(temp_list) + "\n") in_file.close() out_file.close() root@PC1:/home/test3# python test.py root@PC1:/home/test3# ls outcome.ped result.txt test.py root@PC1:/home/test3# paste outcome.ped result.txt ## 结果文件 G G A A T G 2 0 3 1 G G G C T T 0 1 3 2 G G C C T T 0 2 2 2 G G A C G G 1 1 4 0 G G C C G G 0 2 4 0 G G A C T G 1 1 3 1




 浙公网安备 33010602011771号
浙公网安备 33010602011771号