利用基因型数据构建G矩阵
001、plink + sommer实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --recode A 1> /dev/null ## 将A\T\C\G转换为0\1\2形式 root@PC1:/home/test# ls outcome.map outcome.ped plink.log plink.nosex plink.raw
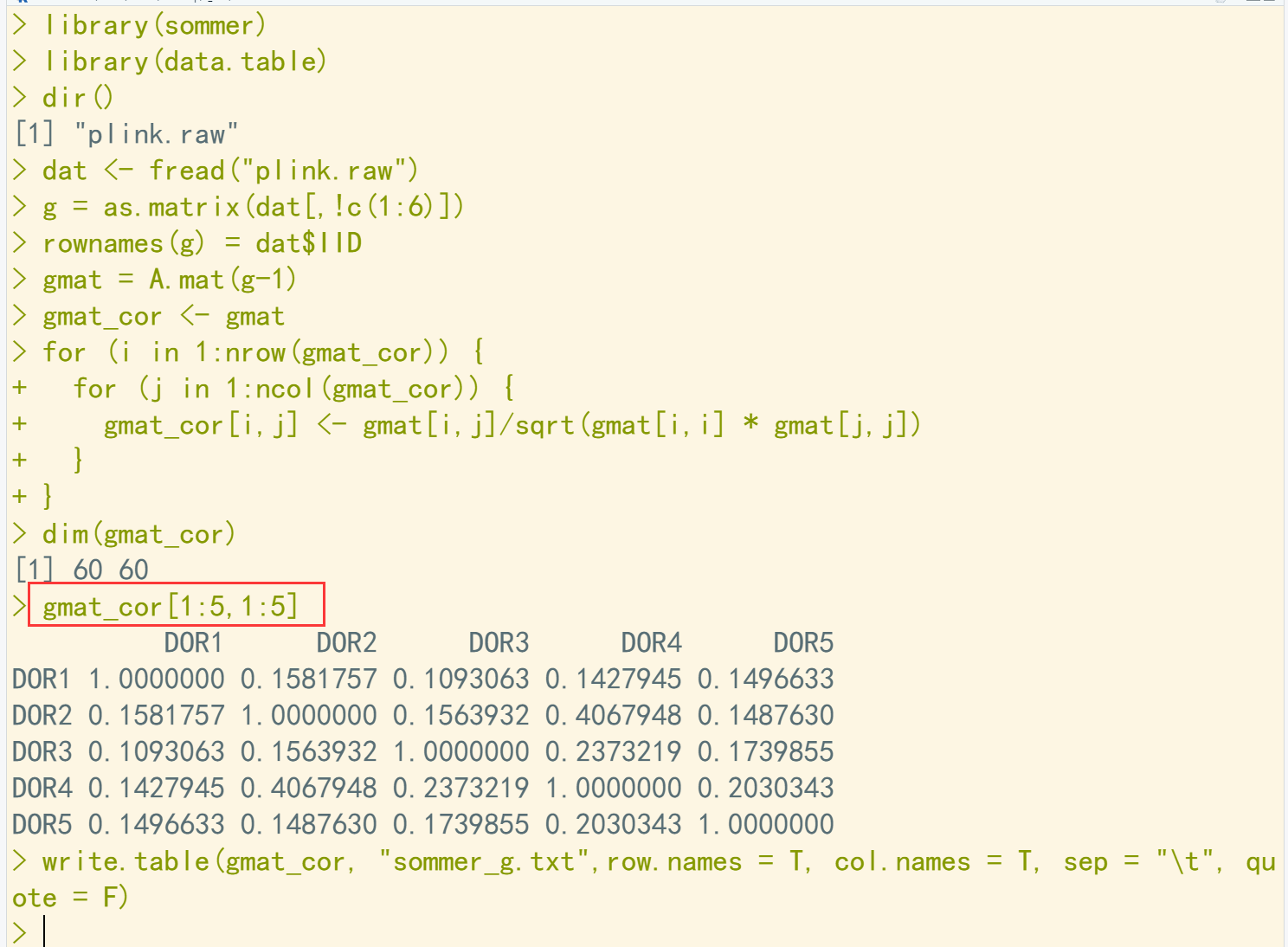
library(sommer) library(data.table) dir() dat <- fread("plink.raw") g = as.matrix(dat[,!c(1:6)]) rownames(g) = dat$IID gmat = A.mat(g-1) gmat_cor <- gmat for (i in 1:nrow(gmat_cor)) { for (j in 1:ncol(gmat_cor)) { gmat_cor[i,j] <- gmat[i,j]/sqrt(gmat[i,i] * gmat[j,j]) } } dim(gmat_cor) gmat_cor[1:5,1:5] write.table(gmat_cor, "sommer_g.txt",row.names = T, col.names = T, sep = "\t", quote = F)
002、gemma实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-bed 1> /dev/null ## 生成二进制文件 root@PC1:/home/test# ls outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# /home/software/gemma-0.98.5-linux-static-AMD64 -bfile plink -gk 2 -o gemma -outdir ./ ## 构建g矩阵 GEMMA 0.98.5 (2021-08-25) by Xiang Zhou, Pjotr Prins and team (C) 2012-2021 Reading Files ... ## number of total individuals = 60 ## number of analyzed individuals = 60 ## number of covariates = 1 ## number of phenotypes = 1 ## number of total SNPs/var = 42629 ## number of analyzed SNPs = 42629 Calculating Relatedness Matrix ... ================================================== 100% **** INFO: Done. root@PC1:/home/test# ls gemma.log.txt gemma.sXX.txt outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# head -n 5 gemma.sXX.txt | cut -f 1-5 0.9890465761 0.1073153377 0.1182919093 0.1263857559 0.1589786895 0.1073153377 0.9722035427 0.1886718834 0.2216020996 0.1799985087 0.1182919093 0.1886718834 0.9714687294 0.1556666191 0.1414708648 0.1263857559 0.2216020996 0.1556666191 0.9272709102 0.141052078 0.1589786895 0.1799985087 0.1414708648 0.141052078 0.9287239597
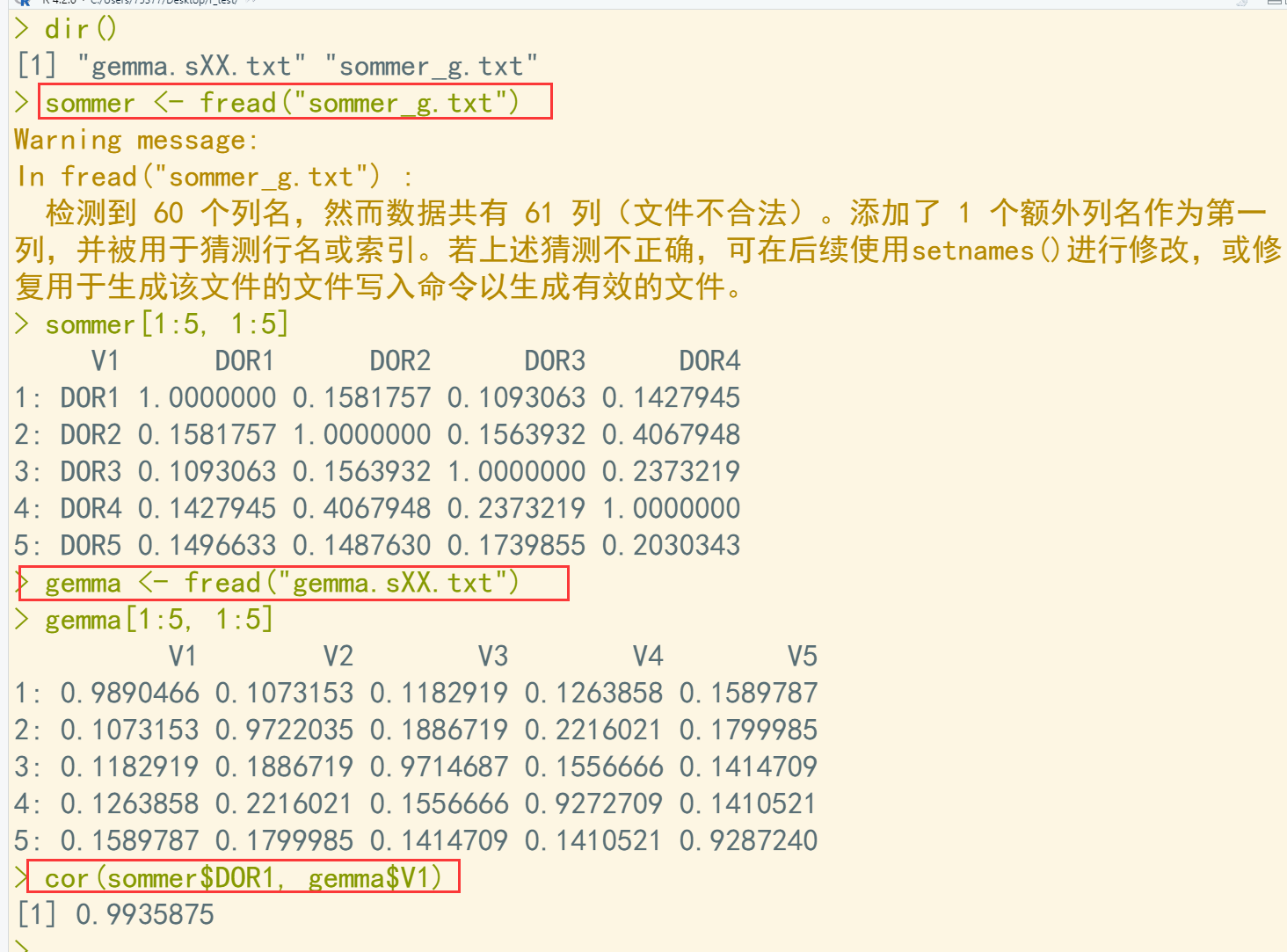
dir() sommer <- fread("sommer_g.txt") sommer[1:5, 1:5] gemma <- fread("gemma.sXX.txt") gemma[1:5, 1:5] cor(sommer$DOR1, gemma$V1) ## 计算相关系数
003、plink实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-rel square 1> /dev/null ## plink生成G矩阵 root@PC1:/home/test# ls outcome.map outcome.ped plink.log plink.rel plink.rel.id root@PC1:/home/test# head -n 6 plink.rel | cut -f 1-6 1.04625 0.122221 0.132869 0.142839 0.178275 0.131377 0.122221 1.03097 0.209148 0.246329 0.201715 0.235886 0.132869 0.209148 1.02611 0.174476 0.160126 0.206312 0.142839 0.246329 0.174476 0.979224 0.16131 0.2332 0.178275 0.201715 0.160126 0.16131 0.98128 0.135685 0.131377 0.235886 0.206312 0.2332 0.135685 0.984521
dir() sommer <- fread("sommer_g.txt") sommer[1:3, 1:3] gemma <- fread("gemma.sXX.txt") gemma[1:3, 1:3] plink <- fread("plink.rel") plink[1:3, 1:3] cor(sommer$DOR1, plink$V1) cor(gemma$V1, plink$V1)

004、gcta实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-bed 1> /dev/null ## 转二进制 root@PC1:/home/test# ls outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log ## 构建矩阵 root@PC1:/home/test# /home/software/gcta_v1.94.0Beta_linux_kernel_3_x86_64/gcta_v1.94.0Beta_linux_kernel_3_x86_64_static --bfile plink --make-grm-gz --make-grm-alg 1 --out gcta 1> /dev/null root@PC1:/home/test# ls gcta.grm.gz gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# gunzip gcta.grm. gzip: gcta.grm..gz: No such file or directory root@PC1:/home/test# gunzip gcta.grm.gz root@PC1:/home/test# ls gcta.grm gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# head -n 3 gcta.grm ## 结果文件 1 1 1.593672e+04 1.068706e+00 2 1 1.593672e+04 1.294735e-01 2 2 1.593672e+04 1.078836e+00 root@PC1:/home/test# cut -f 1,2,4 gcta.grm > gcta.grm2 ## 修改格式 root@PC1:/home/test# ls gcta.grm gcta.grm2 gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log
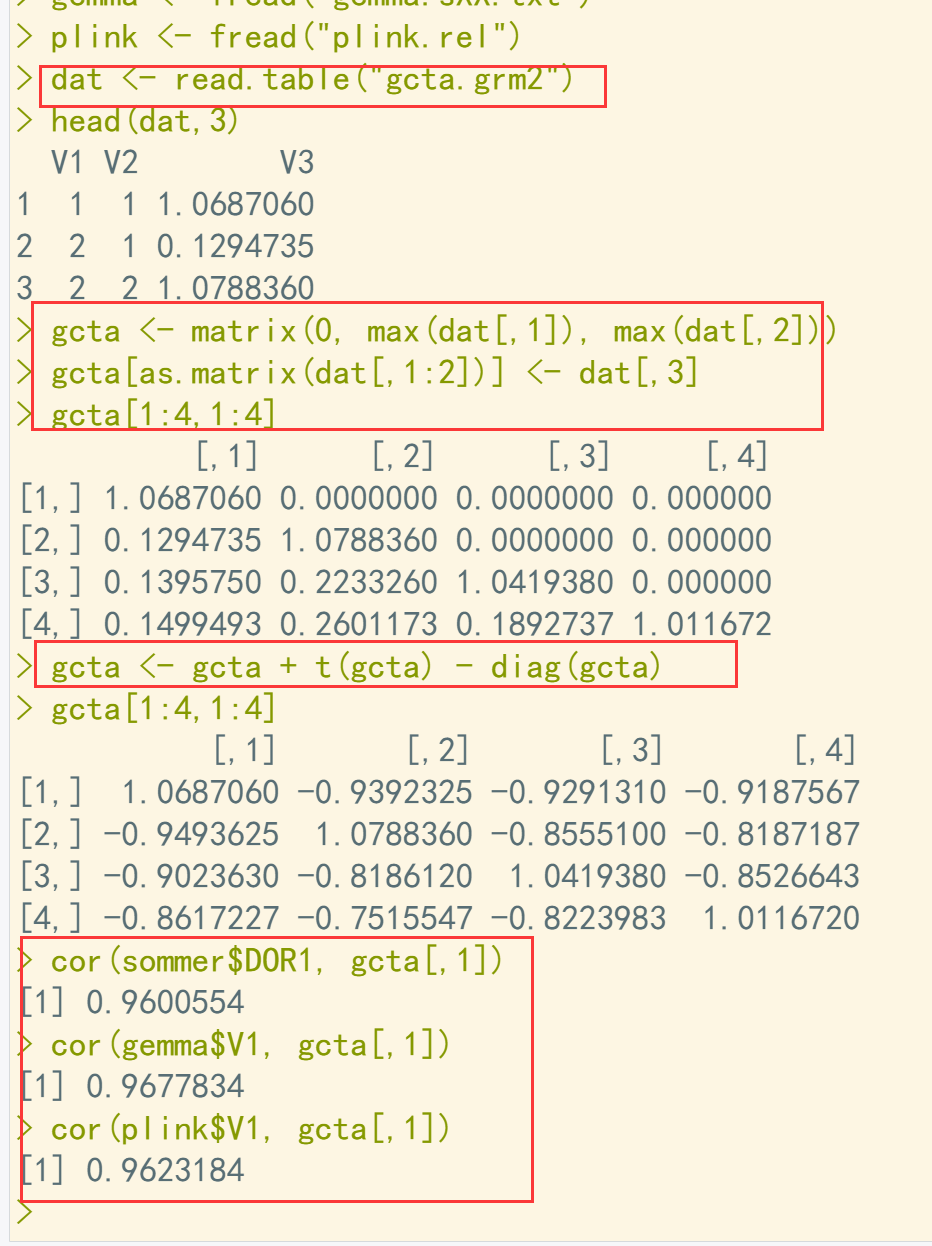
dir() sommer <- fread("sommer_g.txt") gemma <- fread("gemma.sXX.txt") plink <- fread("plink.rel") dat <- read.table("gcta.grm2") head(dat,3) gcta <- matrix(0, max(dat[,1]), max(dat[,2])) gcta[as.matrix(dat[,1:2])] <- dat[,3] ## 三元组转换为矩阵形式 gcta[1:4,1:4] gcta <- gcta + t(gcta) - diag(gcta) gcta[1:4,1:4] cor(sommer$DOR1, gcta[,1]) cor(gemma$V1, gcta[,1]) cor(plink$V1, gcta[,1])




 浙公网安备 33010602011771号
浙公网安备 33010602011771号