GWAS分析中一般线性模型GML + 协变量中snp的回归系数r2的计算
001、plink + R
root@PC1:/home/test# ls gwas_test.map gwas_test.ped root@PC1:/home/test# plink --file gwas_test --recode A 1> /dev/null root@PC1:/home/test# ls gwas_test.map gwas_test.ped plink.log plink.raw root@PC1:/home/test# plink --file gwas_test --pca 3 header 1> /dev/null root@PC1:/home/test# ls gwas_test.map gwas_test.ped plink.eigenval plink.eigenvec plink.log plink.raw root@PC1:/home/test# awk '{print $2,$6}' plink.raw | paste -d " " - <(cut -d " " -f 3- plink.eigenvec ) | paste -d " " - <(cut -d " " -f 7- plink.raw ) > dat.txt root@PC1:/home/test# head -n 3 dat.txt | cut -d " " -f 1-10 | column -t IID PHENOTYPE PC1 PC2 PC3 snp1_G snp2_T snp3_G snp4_T snp5_G id1 580 -0.0102664 0.0442708 -0.0658835 0 1 1 0 1 id2 690 0.0421124 -0.0313769 -0.0705913 0 0 0 0 0
读入R,计算r2:
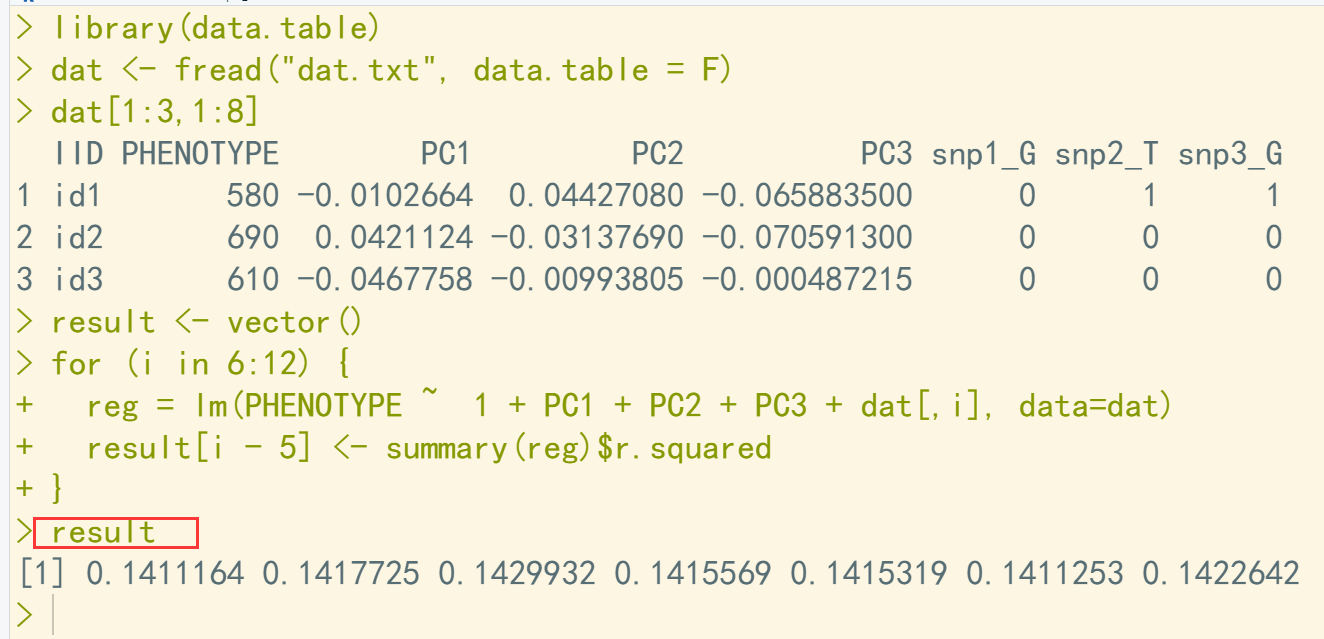
library(data.table) dat <- fread("dat.txt", data.table = F) dat[1:3,1:8] result <- vector() for (i in 6:12) { reg = lm(PHENOTYPE ~ 1 + PC1 + PC2 + PC3 + dat[,i], data=dat) result[i - 5] <- summary(reg)$r.squared } result




 浙公网安备 33010602011771号
浙公网安备 33010602011771号