笔记 4 JM's FSL for task fMRI
Contents
p2 QA #1_ Check your newly created NIfTI files
p3 Brain extraction & QA
p14 Run your level 2 analyses (when you have 3 levels)
p16 group analysis
p2 QA #1_ Check your newly created NIfTI files
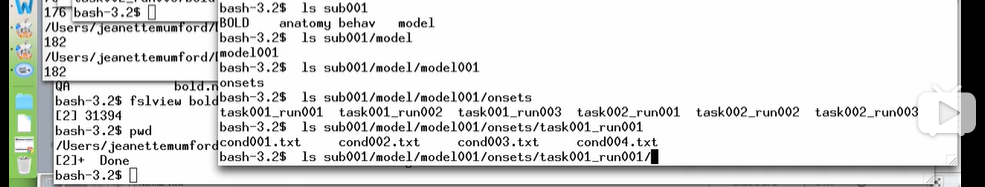
1. 被试数据的层级结构
subjects(?)/subXXX/
/BOLD/
/QA
/taskXXX_runXXX/bold.nii.gz
/anatomy
/behav
/model/modelXXX/onsets/taskXXX_runXXX/conXXX.txt (change according to personal preference)
(https://www.bilibili.com/video/av61429264?p=2,04:40, 08:02, 08:55)
2. 同上图,一个run里用不同的txt文件来设置不同条件/刺激/对比(contrasts)
3. 开始处理前需检查的几点

(https://www.bilibili.com/video/av61429264?p=2,18:12)
*[0-9] to list all numbers @ this position. i.e., ls cond00[0-9].txt to list cond001.txt cond002.txt cond003.txt ... cond009.txt, if any of them exist.
**dim4 listed with fslinfo represents # of volumes
***fslview T1.nii.gz works well to show orientation labels (exactly resemble the following pic)
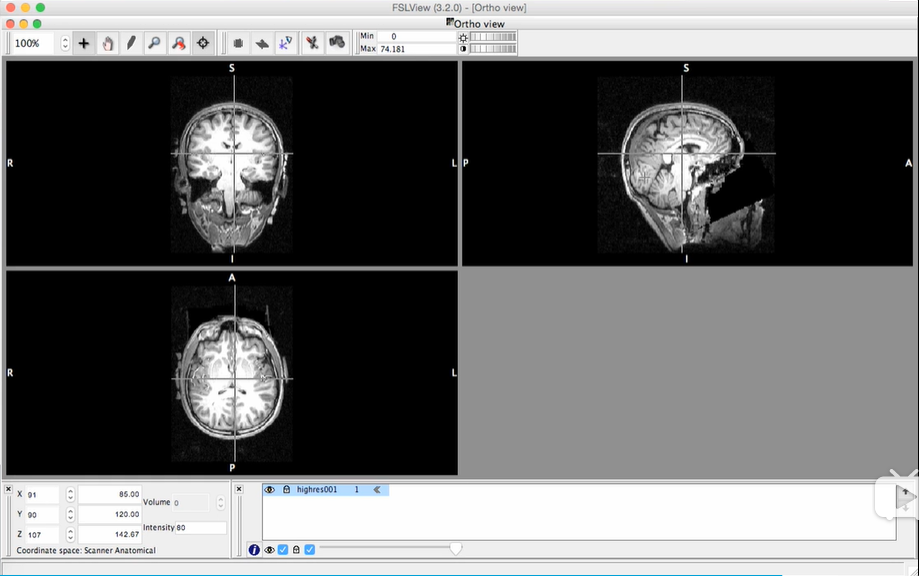
(https://www.bilibili.com/video/av61429264?p=2, 16:01)
p3
1. 对结构像去掉亮部,尤其是眼球和头盖骨(skull)
2. (https://www.bilibili.com/video/av61429264?p=3, 5:14)

-R??
-g !! seems good to use
3. freesurfer 剥脑壳更好(12:00)
1. 分fsf对每个被试的三个run做平均(为什么不把所有被试放进一个fsf,有什么差别?)
2. 10:48 对paired的一种设置方式
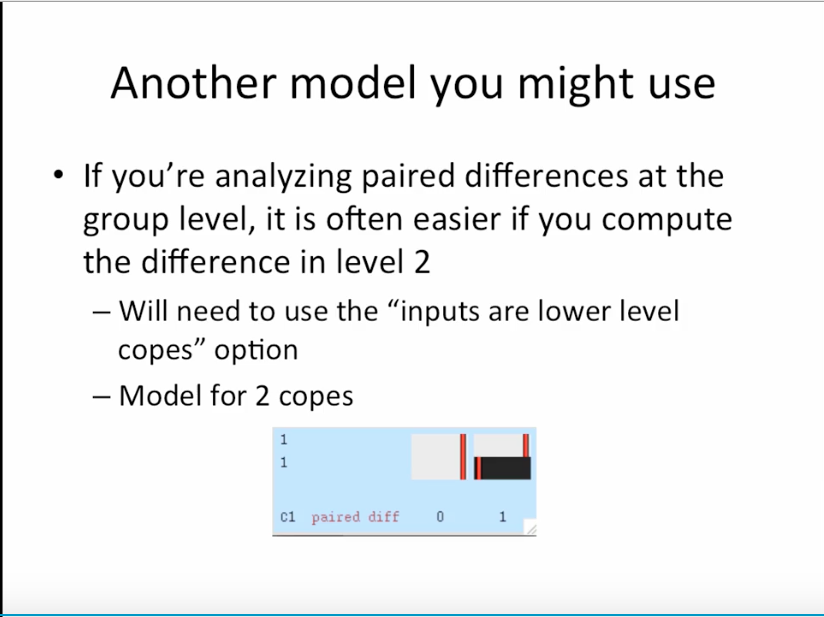
1. do flame1 only at this level and do thresholding like this
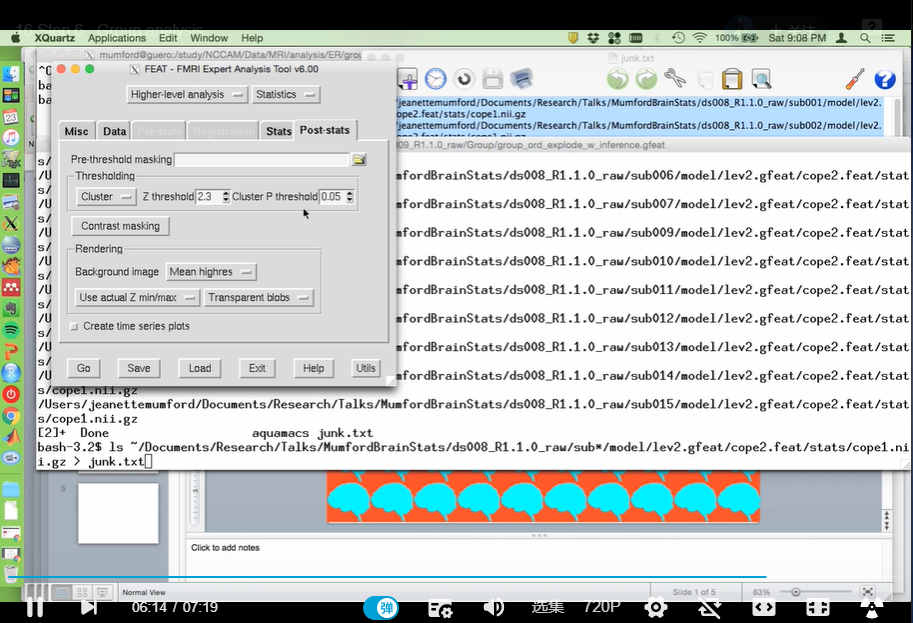
1. thresh_zstat1.nii.gz是经过多重比较校正的文件,stat文件夹里的zstat是未经过校正的(fsleyes时最小值填2)
2. flip through func_filtered_data to check for dependent variable

tbc



 浙公网安备 33010602011771号
浙公网安备 33010602011771号