


















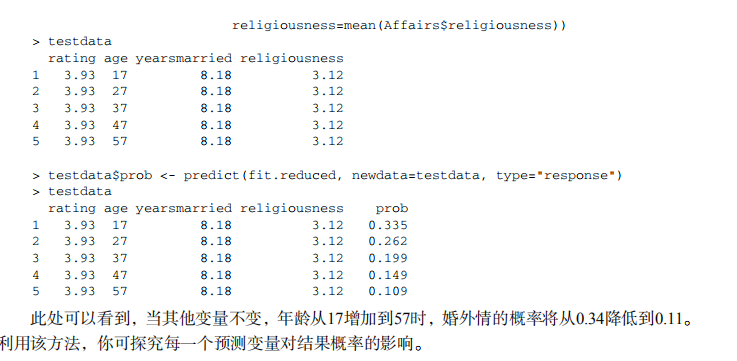



#----------------------------------------------# # R in Action (2nd ed): Chapter 13 # # Generalized linear models # # requires packages AER, robust, gcc # # install.packages(c("AER", "robust", "gcc")) # #----------------------------------------------# ## Logistic Regression # get summary statistics data(Affairs, package="AER") summary(Affairs) table(Affairs$affairs) # create binary outcome variable Affairs$ynaffair[Affairs$affairs > 0] <- 1 Affairs$ynaffair[Affairs$affairs == 0] <- 0 Affairs$ynaffair <- factor(Affairs$ynaffair, levels=c(0,1), labels=c("No","Yes")) table(Affairs$ynaffair) # fit full model fit.full <- glm(ynaffair ~ gender + age + yearsmarried + children + religiousness + education + occupation +rating, data=Affairs,family=binomial()) summary(fit.full) # fit reduced model fit.reduced <- glm(ynaffair ~ age + yearsmarried + religiousness + rating, data=Affairs, family=binomial()) summary(fit.reduced) # compare models anova(fit.reduced, fit.full, test="Chisq") # interpret coefficients coef(fit.reduced) exp(coef(fit.reduced)) # calculate probability of extramariatal affair by marital ratings testdata <- data.frame(rating = c(1, 2, 3, 4, 5), age = mean(Affairs$age), yearsmarried = mean(Affairs$yearsmarried), religiousness = mean(Affairs$religiousness)) testdata$prob <- predict(fit.reduced, newdata=testdata, type="response") testdata # calculate probabilites of extramariatal affair by age testdata <- data.frame(rating = mean(Affairs$rating), age = seq(17, 57, 10), yearsmarried = mean(Affairs$yearsmarried), religiousness = mean(Affairs$religiousness)) testdata$prob <- predict(fit.reduced, newdata=testdata, type="response") testdata # evaluate overdispersion fit <- glm(ynaffair ~ age + yearsmarried + religiousness + rating, family = binomial(), data = Affairs) fit.od <- glm(ynaffair ~ age + yearsmarried + religiousness + rating, family = quasibinomial(), data = Affairs) pchisq(summary(fit.od)$dispersion * fit$df.residual, fit$df.residual, lower = F) ## Poisson Regression # look at dataset data(breslow.dat, package="robust") names(breslow.dat) summary(breslow.dat[c(6, 7, 8, 10)]) # plot distribution of post-treatment seizure counts opar <- par(no.readonly=TRUE) par(mfrow=c(1, 2)) attach(breslow.dat) hist(sumY, breaks=20, xlab="Seizure Count", main="Distribution of Seizures") boxplot(sumY ~ Trt, xlab="Treatment", main="Group Comparisons") par(opar) # fit regression fit <- glm(sumY ~ Base + Age + Trt, data=breslow.dat, family=poisson()) summary(fit) # interpret model parameters coef(fit) exp(coef(fit)) # evaluate overdispersion deviance(fit)/df.residual(fit) library(qcc) qcc.overdispersion.test(breslow.dat$sumY, type="poisson") # fit model with quasipoisson fit.od <- glm(sumY ~ Base + Age + Trt, data=breslow.dat, family=quasipoisson()) summary(fit.od)



