SIFT4G
SIFT4G
SIFT帮助文档SIFT HELP
SIFT官网SIFT HOME
Annotate variants with SIFT4G
SIFT 4G Annotator will annotate a variant list (.vcf file) with predictions from a SIFT 4G database.
! VCF file must be sorted by chromosome and position to be annotated properly.
1. If your organism is not listed, you can create your own SIFT prediction database
2. Annotate variants:
To run the SIFT 4G Annotator on Linux or Mac via command line, type the following command into the terminal:
java -jar <Path to SIFT4G_Annotator> -c -i <Path to input vcf file> -d <Path to SIFT4G database directory> -r <Path to your results folder> -t
Note:To run the Annotator via command line "-c" is essential (see the commandline parameters in the table below). If "-t" option is not used SIFT 4G extracts annotator single transcript per variant.
Command line Options:
| Option | Description |
|---|---|
| -c | To run on command line |
| -i | Path to your input variants file in VCF format |
| -d | Path to SIFT database directory |
| -r | Path to your output results folder |
| -t | To extract annotations for multiple transcripts (Optional) |
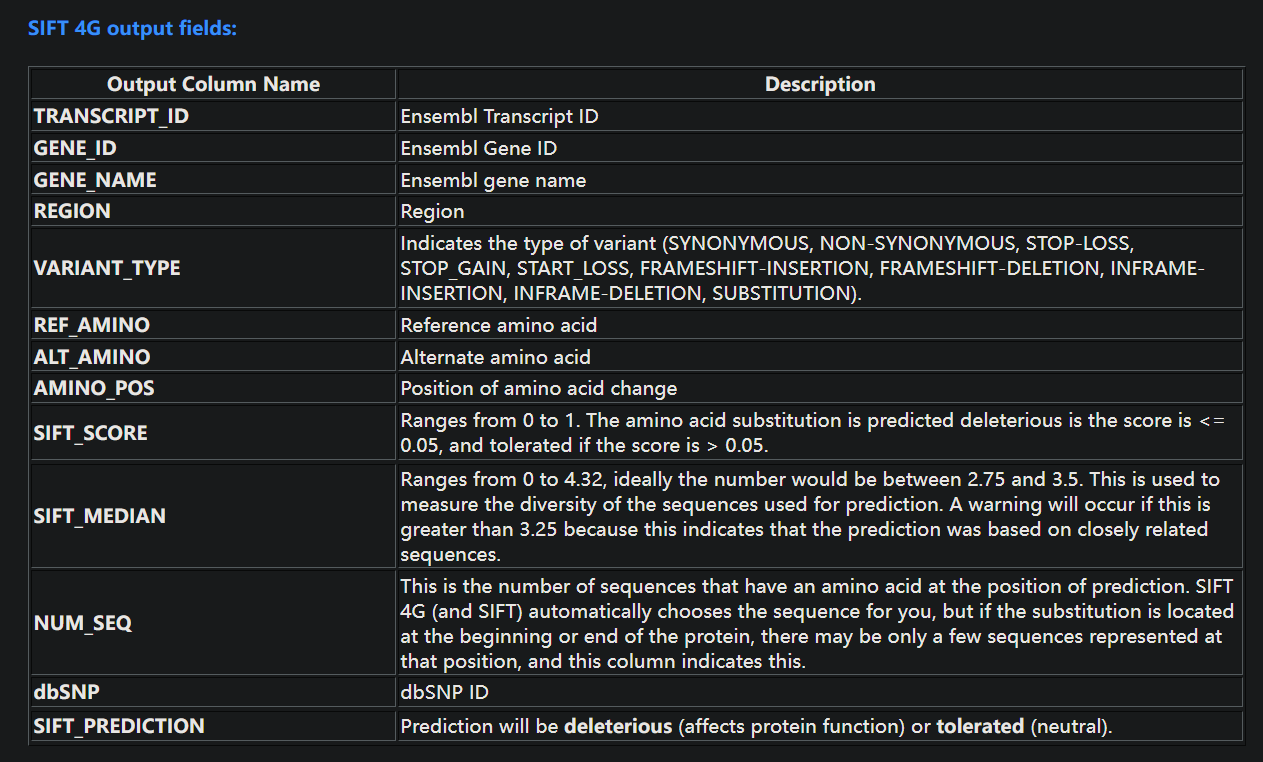
3. SIFT 4G Output
| CHROM | POSITION | REF_ALLELE | ALT_ALLELE | TRANSCRIPT_ID | GENE_ID | GENE_NAME | REGION | VARIANT_TYPE | REF_AA | ALT_AA | AA_POS | SIFT_SCORE | SIFT_MEDIAN | NUM_SEQs | dbSNP | PREDICTION |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 881918 | G | A | ENST00000327044 | ENSG00000188976 | NOC2L | CDS | NONSYNONYMOUS | S | L | 556 | 0.095 | 2.54 | 44 | rs35471880 | TOLERATED |
| 1 | 900505 | G | C | ENST00000338591 | ENSG00000187961 | KLHL17 | CDS | SYNONYMOUS | V | V | 621 | 1 | 2.62 | 79 | rs28705211 | TOLERATED |
| 1 | 900717 | CTTAT | C | ENST00000338591 | ENSG00000187961 | KLHL17 | UTR_3 | FRAMESHIFT | DELETION | NA | NA | NA | NA | NA | NA | novel |




 浙公网安备 33010602011771号
浙公网安备 33010602011771号