ResearchSphere: End-to-End Document Research
ResearchSphere: End-to-End Document Research
https://github.com/SaiPranaviJeedigunta/Multi-Agent-RAG-Application/tree/main
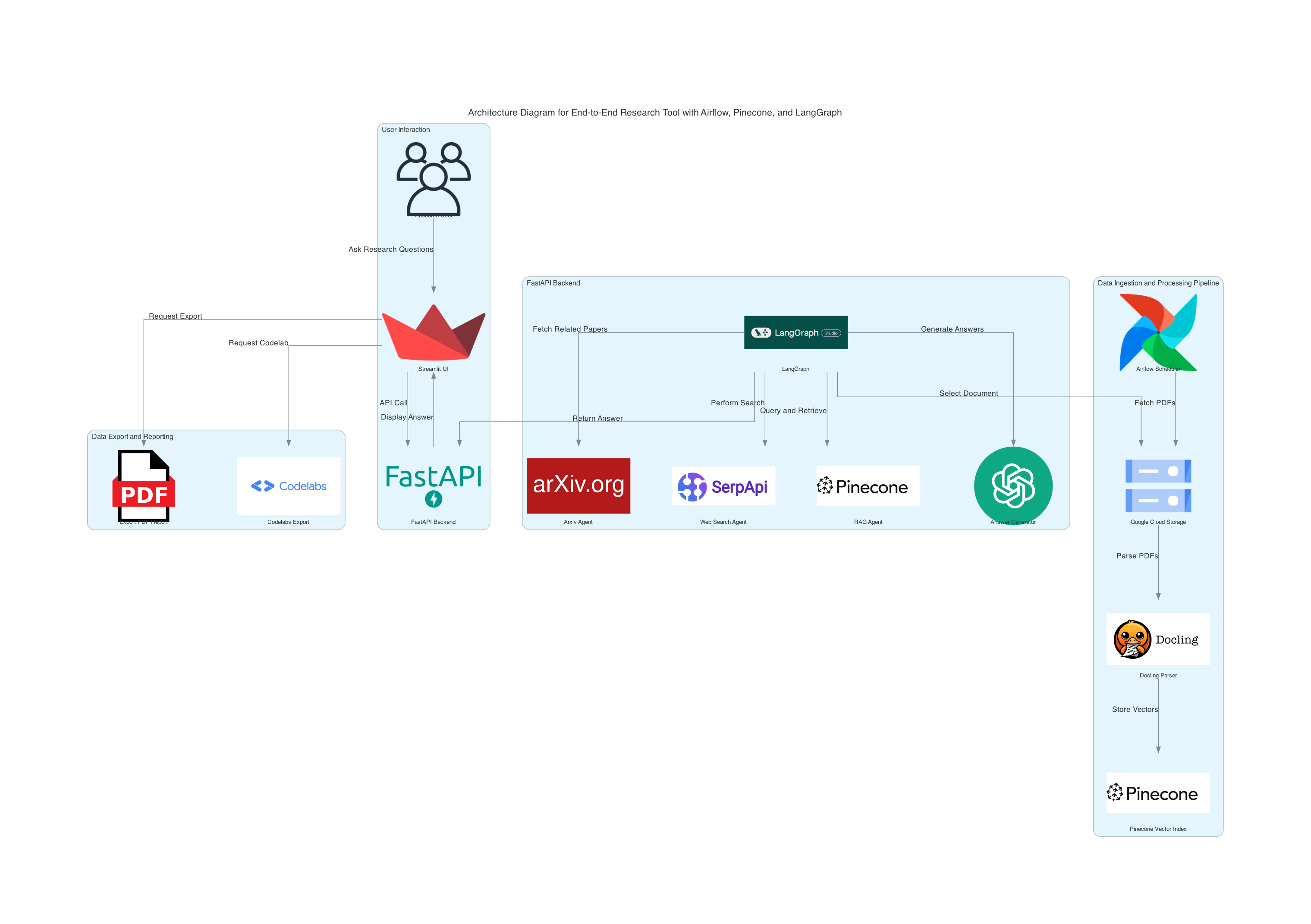
Build an end-to-end research tool using an Airflow pipeline for document processing, vector storage with Pinecone, and a multi-agent research system powered by LangGraph. The system enables efficient document-based research, query answering, and exportable findings.
Research often involves parsing through extensive documents, retrieving relevant data, and synthesizing actionable insights. Current methods are time-consuming and inefficient, relying heavily on manual work. This project addresses the need for automation and intelligence in research workflows by building a pipeline to process documents, store vectors for similarity search, and create a multi-agent system for enhanced research capabilities.
This project automates the ingestion, processing, and retrieval of publication data using advanced technologies. Key features include:
- Airflow Pipeline: Automates document parsing using Docling and stores vectors in Pinecone for similarity search.
- Multi-Agent System: Utilizes LangGraph to orchestrate research tasks, including document selection, Arxiv-based research, web search, and answering queries with Retrieval-Augmented Generation (RAG).
- User Interaction: Provides an interface using Streamlit for users to ask questions, save research sessions, and export results in professional formats.
- Airflow Pipeline:
- Uses Docling to parse documents, extracting structured text from PDFs.
- Stores document vectors in Pinecone for efficient similarity search.
- Automates the end-to-end pipeline using Apache Airflow.
- Technologies:
- Docling for document parsing.
- Pinecone for vector-based storage and retrieval.
- Airflow for automation and orchestration.
-
Research Agents:
- Document Selector: Enables selection of documents processed in Part 1.
- Arxiv Agent: Searches for related research papers on Arxiv.
- Web Search Agent: Performs broader online research for context.
- RAG Agent: Uses Retrieval-Augmented Generation (RAG) to answer questions based on the document's content.
- Answer Generator: Provides concise answers or summaries using OpenAI or similar APIs.
-
Workflow:
- Agents are orchestrated by LangGraph, interacting with Pinecone for vector retrieval and external APIs for additional insights.
- Pinecone facilitates fast similarity search, enhancing query efficiency.
- User Interface:
- Built using Streamlit to allow users to:
- Ask 5-6 questions per document.
- View structured answers or summaries.
- Save research session results.
- Built using Streamlit to allow users to:
- Export Results:
- PDF Export: Generates professional reports summarizing findings.
- Codelabs Export: Structures findings for instructional use.
- Document Processing:
- Automates document ingestion, parsing, and vector storage.
- Multi-Agent System:
- Offers advanced research capabilities through document selection, Arxiv search, web search, and RAG-based Q/A.
- User-Friendly Interface:
- Simple yet powerful interface for research interaction and result export.
DATA INGESTIONN
https://github.com/SaiPranaviJeedigunta/Multi-Agent-RAG-Application/blob/main/airflow/dags/datapipeline.py
from google.cloud import storage from dotenv import load_dotenv import os from google.oauth2 import service_account from google.cloud import storage from airflow import DAG from airflow.operators.python_operator import PythonOperator from datetime import datetime load_dotenv() # Get the path to the credentials file from environment variable credentials_path = os.getenv("GOOGLE_APPLICATION_CREDENTIALS") # Create credentials and client if credentials_path: credentials = service_account.Credentials.from_service_account_file(credentials_path) client = storage.Client(credentials=credentials) else: raise ValueError("GOOGLE_APPLICATION_CREDENTIALS environment variable not set") def download_files_from_gcs(): # Fetch bucket name from environment variable bucket_name = os.getenv("GCS_BUCKET_NAME") # Check if bucket name is loaded correctly if not bucket_name: raise ValueError("Bucket name not found. Ensure GCS_BUCKET_NAME is set in .env file.") print("Bucket name:", bucket_name) # Initialize the Google Cloud Storage client client = storage.Client() bucket = client.bucket(bucket_name) # List of specific files to download file_paths = [ "cfai_publications/The Economics of Private Equity: A Critical Review/The Economics of Private Equity: A Critical Review.pdf", "cfai_publications/Investment Horizon, Serial Correlation, and Better (Retirement) Portfolios/Investment Horizon, Serial Correlation, and Better (Retirement) Portfolios.pdf", "cfai_publications/An Introduction to Alternative Credit/An Introduction to Alternative Credit.pdf" ] # Set the current directory as the destination for downloads current_directory = os.getcwd() # Download each file for file_path in file_paths: blob = bucket.blob(file_path) local_file_path = os.path.join(current_directory, os.path.basename(file_path)) blob.download_to_filename(local_file_path) print(f"Downloaded {file_path} to {local_file_path}") def process_and_save_pdfs(): import logging import time from pathlib import Path import pandas as pd import json from docling.datamodel.base_models import InputFormat from docling.datamodel.pipeline_options import PdfPipelineOptions from docling.document_converter import DocumentConverter, PdfFormatOption from docling_core.types.doc import ImageRefMode, PictureItem, TableItem from docling_core.transforms.chunker import HierarchicalChunker # Configure logging logging.basicConfig(level=logging.INFO) _log = logging.getLogger(__name__) # Define the resolution scale for images IMAGE_RESOLUTION_SCALE = 2.0 # Paths to local PDF files file_paths = [ "The Economics of Private Equity: A Critical Review.pdf", "Investment Horizon, Serial Correlation, and Better (Retirement) Portfolios.pdf", "An Introduction to Alternative Credit.pdf" ] # Output directory output_dir = Path("parsed_content") # Replace with your desired output directory path output_dir.mkdir(parents=True, exist_ok=True) # Function to process PDF files def process_pdf(file_path): pipeline_options = PdfPipelineOptions() pipeline_options.images_scale = IMAGE_RESOLUTION_SCALE pipeline_options.generate_page_images = True pipeline_options.generate_table_images = True pipeline_options.generate_picture_images = True doc_converter = DocumentConverter( format_options={ InputFormat.PDF: PdfFormatOption(pipeline_options=pipeline_options) } ) start_time = time.time() conv_res = doc_converter.convert(file_path) doc_filename = Path(file_path).stem # Save page images for page_no, page in conv_res.document.pages.items(): page_image_filename = output_dir / f"{doc_filename}-page-{page_no}.png" with page_image_filename.open("wb") as fp: page.image.pil_image.save(fp, format="PNG") _log.info(f"Saved page image: {page_image_filename}") # Save images of figures and tables table_counter = 0 picture_counter = 0 for element, _level in conv_res.document.iterate_items(): if isinstance(element, TableItem): table_counter += 1 table_image_filename = output_dir / f"{doc_filename}-table-{table_counter}.png" with page_image_filename.open("wb") as fp: element.image.pil_image.save(fp, "PNG") _log.info(f"Saved table image: {table_image_filename}") # Save the table as CSV and HTML table_df: pd.DataFrame = element.export_to_dataframe() table_csv_filename = output_dir / f"{doc_filename}-table-{table_counter}.csv" table_df.to_csv(table_csv_filename) table_html_filename = output_dir / f"{doc_filename}-table-{table_counter}.html" with table_html_filename.open("w") as fp: fp.write(element.export_to_html()) _log.info(f"Saved table CSV: {table_csv_filename} and HTML: {table_html_filename}") if isinstance(element, PictureItem): picture_counter += 1 picture_image_filename = output_dir / f"{doc_filename}-picture-{picture_counter}.png" with picture_image_filename.open("wb") as fp: element.image.pil_image.save(fp, "PNG") _log.info(f"Saved picture image: {picture_image_filename}") # Apply hierarchy-aware chunking for further processing chunks = list(HierarchicalChunker(min_chunk_length=500, max_chunk_length=1500, split_by='paragraph', overlap=50).chunk(conv_res.document)) # Prepare to save chunk data chunk_data = [] # Process each chunk and display metadata for i, chunk in enumerate(chunks): text_content = chunk.text # Directly access 'text' attribute # Convert meta information to a dictionary, or extract relevant fields to avoid serialization issues meta_info = chunk.meta.dict() if hasattr(chunk.meta, "dict") else str(chunk.meta) # Store each chunk's content and metadata for further use chunk_metadata = { "document": doc_filename, "chunk_id": i, "text": text_content, "meta": meta_info } chunk_data.append(chunk_metadata) print(f"Chunk {i}: {chunk_metadata['text'][:100]}...") # Preview the first 100 characters # Save chunks data to a JSON file for each document chunks_json_filename = output_dir / f"{doc_filename}_chunks.json" with chunks_json_filename.open("w") as json_fp: json.dump(chunk_data, json_fp, indent=4) _log.info(f"Chunks saved to JSON file: {chunks_json_filename}") # Export markdown with embedded images for content content_md = conv_res.document.export_to_markdown(image_mode=ImageRefMode.EMBEDDED) md_filename = output_dir / f"{doc_filename}-with-images.md" with md_filename.open("w") as fp: fp.write(content_md) _log.info(f"Markdown with images saved: {md_filename}") end_time = time.time() - start_time _log.info(f"{doc_filename} converted and figures exported in {end_time:.2f} seconds.") # Process each file in the list for file_path in file_paths: process_pdf(file_path) def process_and_upload_to_pinecone(): import os import json import glob import logging import pandas as pd from dotenv import load_dotenv from pathlib import Path from pinecone import Pinecone, ServerlessSpec from sentence_transformers import SentenceTransformer from transformers import CLIPProcessor, CLIPModel from PIL import Image import pytesseract import torch import platform os.environ["TOKENIZERS_PARALLELISM"] = "false" # Set the Tesseract path from the environment variable tesseract_path = os.getenv("TESSERACT_PATH") if tesseract_path: pytesseract.pytesseract.tesseract_cmd = tesseract_path else: raise EnvironmentError("TESSERACT_PATH is not set in .env") # Set up logging logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s') # Load environment variables load_dotenv() # Initialize Pinecone with environment variables pinecone_api_key = os.getenv("PINECONE_API_KEY") pinecone_env = "us-east-1" pc = Pinecone(api_key=pinecone_api_key) # Define Pinecone index parameters index_name = "research-publications-index" embedding_model = SentenceTransformer('all-MiniLM-L6-v2') embedding_dimension = embedding_model.get_sentence_embedding_dimension() # Initialize CLIP model and processor for image embeddings clip_model = CLIPModel.from_pretrained("openai/clip-vit-base-patch32") clip_processor = CLIPProcessor.from_pretrained("openai/clip-vit-base-patch32") clip_embedding_dim = clip_model.config.projection_dim # Check if index exists, delete if it does if index_name in pc.list_indexes().names(): logging.info(f"Index '{index_name}' already exists. Deleting the existing index.") pc.delete_index(index_name) # Create the Pinecone index logging.info(f"Creating Pinecone index: {index_name}") pc.create_index( name=index_name, dimension=embedding_dimension, metric='cosine', spec=ServerlessSpec(cloud='aws', region=pinecone_env) ) # Connect to the index index = pc.Index(index_name) logging.info(f"Connected to Pinecone index: {index_name}") # Path to the directory with parsed content parsed_content_dir = "parsed_content" # Function to chunk text into smaller parts if it exceeds max token length def chunk_text(text, max_length=512): return [text[i:i+max_length] for i in range(0, len(text), max_length)] # Step 1: Process each JSON file containing chunked text data for json_file_path in glob.glob(os.path.join(parsed_content_dir, "*_chunks.json")): with open(json_file_path, 'r') as f: chunks = json.load(f) for chunk in chunks: try: # Extract text and metadata text_content = chunk['text'] pdf_filename = chunk['meta']['origin']['filename'] page_no = chunk['meta']['doc_items'][0]['prov'][0].get("page_no") # Chunk large text content for text_chunk in chunk_text(text_content): embedding = embedding_model.encode(text_chunk).tolist() # Metadata for each chunk metadata = { "document": chunk['document'], "chunk_id": chunk['chunk_id'], "page_no": page_no, "type": "text_chunk", "pdf_filename": pdf_filename } # Upload the chunk to Pinecone index.upsert([(f"{chunk['document']}_{chunk['chunk_id']}", embedding, metadata)]) logging.info(f"Uploaded text chunk {chunk['chunk_id']} from document '{chunk['document']}'") except Exception as e: logging.error(f"Failed to process text chunk in {json_file_path}: {e}") # Step 2: Embed and upload each table row with pdf_filename reference for table_csv_path in glob.glob(os.path.join(parsed_content_dir, "*-table-*.csv")): try: doc_filename = Path(table_csv_path).stem.split('-table')[0] # Read the table data table_data = pd.read_csv(table_csv_path) # Iterate over each row in the table for row_idx, row in table_data.iterrows(): row_data = row.to_string() row_embedding = embedding_model.encode(row_data).tolist() # Metadata for each row row_metadata = { "document": doc_filename, "type": "table_row", "row_index": row_idx, "filename": os.path.basename(table_csv_path), "pdf_filename": f"{doc_filename}.pdf" } # Upsert row data into Pinecone index.upsert([(f"{doc_filename}_table_row_{row_idx}", row_embedding, row_metadata)]) logging.info(f"Uploaded row {row_idx} of table from '{table_csv_path}'") except Exception as e: logging.error(f"Failed to process table file {table_csv_path}: {e}") # Step 3: Extract text from images, embed, and upload to Pinecone for image_path in glob.glob(os.path.join(parsed_content_dir, "*-page-*.png")) + glob.glob(os.path.join(parsed_content_dir, "*-picture-*.png")): try: doc_filename = Path(image_path).stem.split('-page')[0].split('-picture')[0] # Load and preprocess image for CLIP image = Image.open(image_path) # Extract text from image using OCR extracted_text = pytesseract.image_to_string(image) logging.info(f"Extracted text from image '{image_path}': {extracted_text[:100]}...") # Embed extracted text using the embedding model if extracted_text.strip(): text_embedding = embedding_model.encode(extracted_text).tolist() text_metadata = { "document": doc_filename, "type": "image_text", "filename": os.path.basename(image_path), "pdf_filename": f"{doc_filename}.pdf" } # Upload extracted text embedding to Pinecone index.upsert([(f"{doc_filename}_{Path(image_path).stem}_text", text_embedding, text_metadata)]) logging.info(f"Uploaded extracted text from image '{image_path}'") # Create image embedding with CLIP inputs = clip_processor(images=image, return_tensors="pt") with torch.no_grad(): image_embedding = clip_model.get_image_features(**inputs).squeeze().tolist() # Truncate the image embedding to match text embedding dimension truncated_image_embedding = image_embedding[:embedding_dimension] # Image metadata image_metadata = { "document": doc_filename, "type": "image", "filename": os.path.basename(image_path), "pdf_filename": f"{doc_filename}.pdf" } # Upload image data into Pinecone index.upsert([(f"{doc_filename}_{Path(image_path).stem}", truncated_image_embedding, image_metadata)]) logging.info(f"Uploaded image data for '{image_path}'") except Exception as e: logging.error(f"Failed to process image file {image_path}: {e}") logging.info("Data successfully embedded and uploaded to Pinecone.") # Define the DAG with DAG( dag_id='parse_and_pinecone_upload', schedule_interval='@daily', # Set the desired schedule interval start_date=datetime(2024, 11, 12), # Change to your desired start date catchup=False, tags=['parsing', 'pineconeupload'] ) as dag: download_publications = PythonOperator( task_id='download_publications', python_callable=download_files_from_gcs ) parse_task = PythonOperator( task_id='parse_task', python_callable=process_and_save_pdfs ) pineconeupload_task = PythonOperator( task_id='pineconeupload_task', python_callable=process_and_upload_to_pinecone ) # Set task dependencies download_publications >> parse_task >> pineconeupload_task
DAG映射
x-airflow-common:
&airflow-common
# In order to add custom dependencies or upgrade provider packages you can use your extended image.
# Comment the image line, place your Dockerfile in the directory where you placed the docker-compose.yaml
# and uncomment the "build" line below, Then run `docker-compose build` to build the images.
image: ${AIRFLOW_IMAGE_NAME:-apache/airflow:2.10.3}
# build: .
environment:
&airflow-common-env
AIRFLOW__CORE__EXECUTOR: CeleryExecutor
AIRFLOW__DATABASE__SQL_ALCHEMY_CONN: postgresql+psycopg2://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__RESULT_BACKEND: db+postgresql://airflow:airflow@postgres/airflow
AIRFLOW__CELERY__BROKER_URL: redis://:@redis:6379/0
AIRFLOW__CORE__FERNET_KEY: ''
AIRFLOW__CORE__DAGS_ARE_PAUSED_AT_CREATION: 'true'
AIRFLOW__CORE__LOAD_EXAMPLES: 'false'
AIRFLOW__API__AUTH_BACKENDS: 'airflow.api.auth.backend.basic_auth,airflow.api.auth.backend.session'
# yamllint disable rule:line-length
# Use simple http server on scheduler for health checks
# See https://airflow.apache.org/docs/apache-airflow/stable/administration-and-deployment/logging-monitoring/check-health.html#scheduler-health-check-server
# yamllint enable rule:line-length
AIRFLOW__SCHEDULER__ENABLE_HEALTH_CHECK: 'true'
# WARNING: Use _PIP_ADDITIONAL_REQUIREMENTS option ONLY for a quick checks
# for other purpose (development, test and especially production usage) build/extend Airflow image.
_PIP_ADDITIONAL_REQUIREMENTS: ${_PIP_ADDITIONAL_REQUIREMENTS:- docling pinecone-client sentence-transformers python-dotenv google-cloud-storage apache-airflow}
# The following line can be used to set a custom config file, stored in the local config folder
# If you want to use it, outcomment it and replace airflow.cfg with the name of your config file
# AIRFLOW_CONFIG: '/opt/airflow/config/airflow.cfg'
volumes:
- ${AIRFLOW_PROJ_DIR:-.}/dags:/opt/airflow/dags
- ${AIRFLOW_PROJ_DIR:-.}/logs:/opt/airflow/logs
- ${AIRFLOW_PROJ_DIR:-.}/config:/opt/airflow/config
- ${AIRFLOW_PROJ_DIR:-.}/plugins:/opt/airflow/plugins
- /Users/pranavijs/Desktop/BD4/.env:/opt/airflow/.env
- /Users/pranavijs/Desktop/BD4/project3-439417-441ae24dff81.json:/opt/airflow/project3-439417-441ae24dff81.json
user: "${AIRFLOW_UID:-50000}:0"
depends_on:
&airflow-common-depends-on
redis:
condition: service_healthy
postgres:
condition: service_healthy













 浙公网安备 33010602011771号
浙公网安备 33010602011771号