llama-factory fine-tuning 3 (Dec 13, 2023)
1 Introduction
In this blog, we will use 3 dataset to fine-tuning our model using llama-factory.
2 dataset preparation
2.1 MedQA dataset (address)
The source of the dataset is designed to examine the doctors' professional capability and thus contains a significant number of questions that require multi-hop logical reasoning.
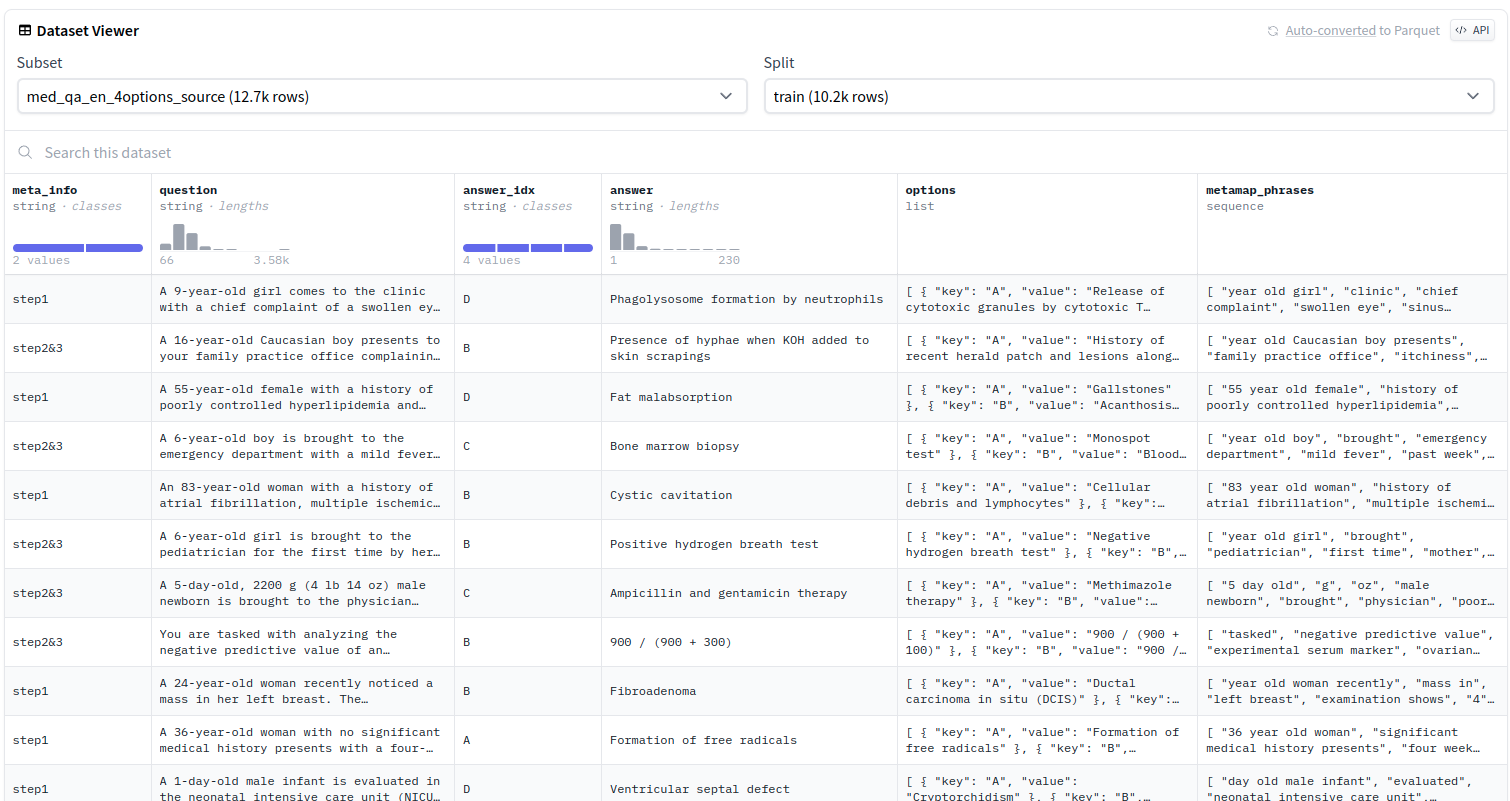
in this dataset, we select question, answer to build our dataset, you can check the following managament code.
click to view the code
from datasets import load_dataset
import os
import json
# Load the dataset
dataset = load_dataset("bigbio/med_qa", "med_qa_en_4options_source")
# Define the save path
save_path = "../medical/MedQA" # Change this path to your local directory
os.makedirs(save_path, exist_ok=True)
# Function to save data as JSON with specified columns
def save_as_json(data, filename):
file_path = os.path.join(save_path, filename)
# Modify the data to include only 'question' and 'answer' columns
data_to_save = [{
"instruction": "Assuming you are a doctor, answer questions based on the patient's symptoms.",
"input": item['question'],
"output": item['answer']
} for item in data]
# Write the modified data to a JSON file
with open(file_path, 'w', encoding='utf-8') as f:
json.dump(data_to_save, f, ensure_ascii=False, indent=4)
# Save the modified data for train, validation, and test splits
save_as_json(dataset['train'], 'train.json')
save_as_json(dataset['validation'], 'validation.json')
save_as_json(dataset['test'], 'test.json')
dataset format
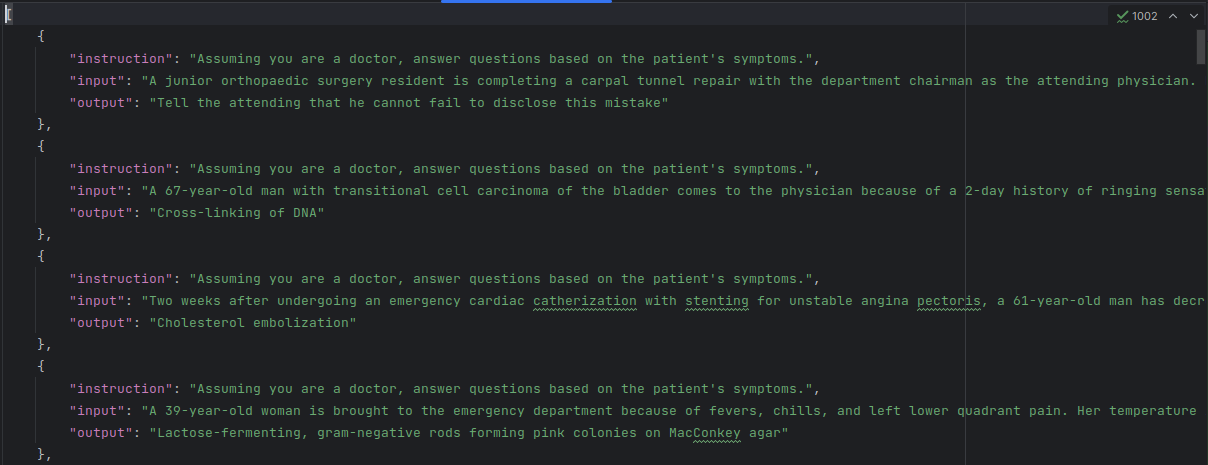
Then we move the dataset (train.json) into llama-factory/data & change it's name as alpaca_med_qa_en.json, then add the following code to llama-factory/data/dataset_info.json
click to view the code
"alpaca_med_qa_en": {
"file_name": "alpaca_med_qa_en.json",
"file_sha1": ""
},
2.2 MedCQA dataset(address)
intorduction: same as MedQA
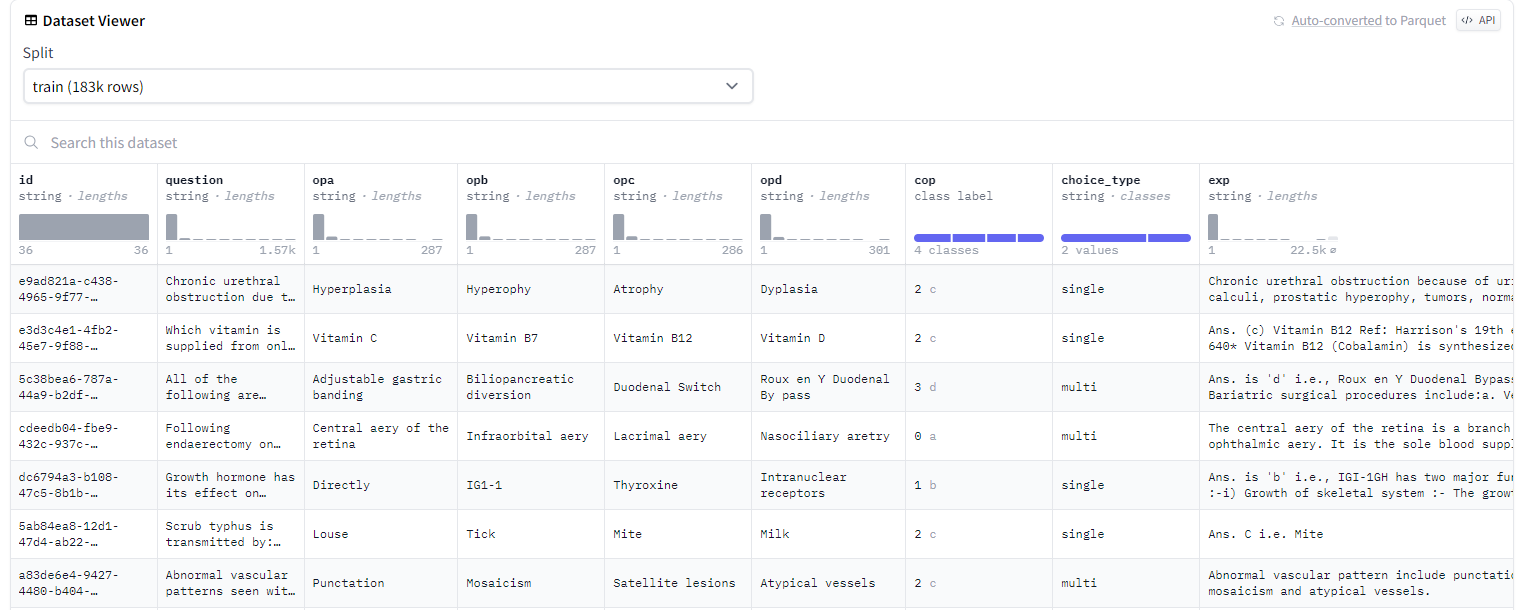
In this dataset, we select question(input), answer+explain(output),for introduction part, we set as "Assuming you are a doctor, answer questions based on the patient's symptoms."
click to view the code
from datasets import load_dataset
import os
import json
# Load the dataset
dataset = load_dataset("medmcqa")
# Define the save path
save_path = "../medical/MedCQA" # Change this path to your local directory
os.makedirs(save_path, exist_ok=True)
# Function to save data as JSON with specified columns
def save_as_json(data, filename):
file_path = os.path.join(save_path, filename)
# Modify the data to include only 'question' and 'answer' columns
data_to_save = [{
"instruction": "Assuming you are a doctor, answer questions based on the patient's symptoms.",
"input": item['question'],
"output": (item['opa'] if item['cop'] == 0 else
item['opb'] if item['cop'] == 1 else
item['opc'] if item['cop'] == 2 else
item['opd'] if item['cop'] == 3 else "No valid answer found") + ": " + (item['exp'] if item['exp'] is not None else "No explanation provided")
} for item in data]
# Write the modified data to a JSON file
with open(file_path, 'w', encoding='utf-8') as f:
json.dump(data_to_save, f, ensure_ascii=False, indent=4)
# Save the modified data for train, validation, and test splits
save_as_json(dataset['train'], 'train.json')
save_as_json(dataset['validation'], 'validation.json')
save_as_json(dataset['test'], 'test.json')
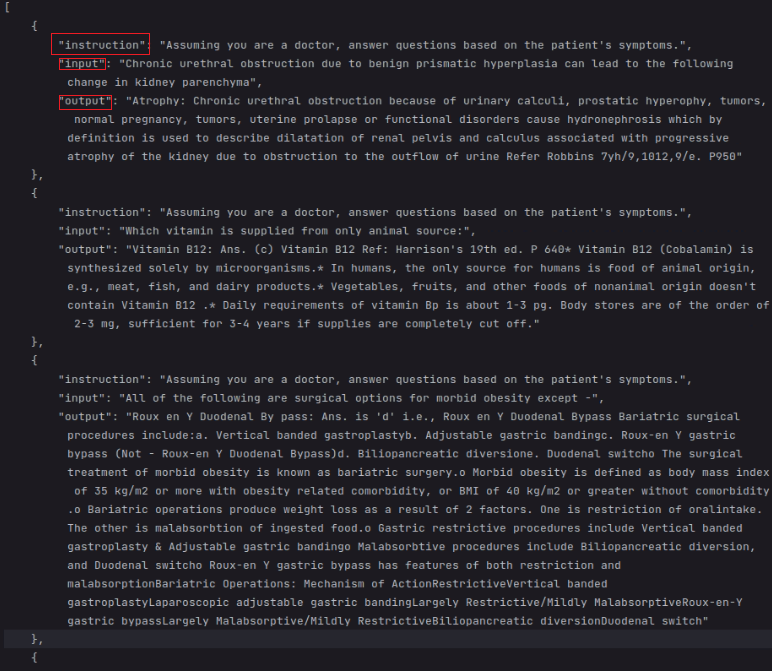
2.3 PubMedQA dataset (address)
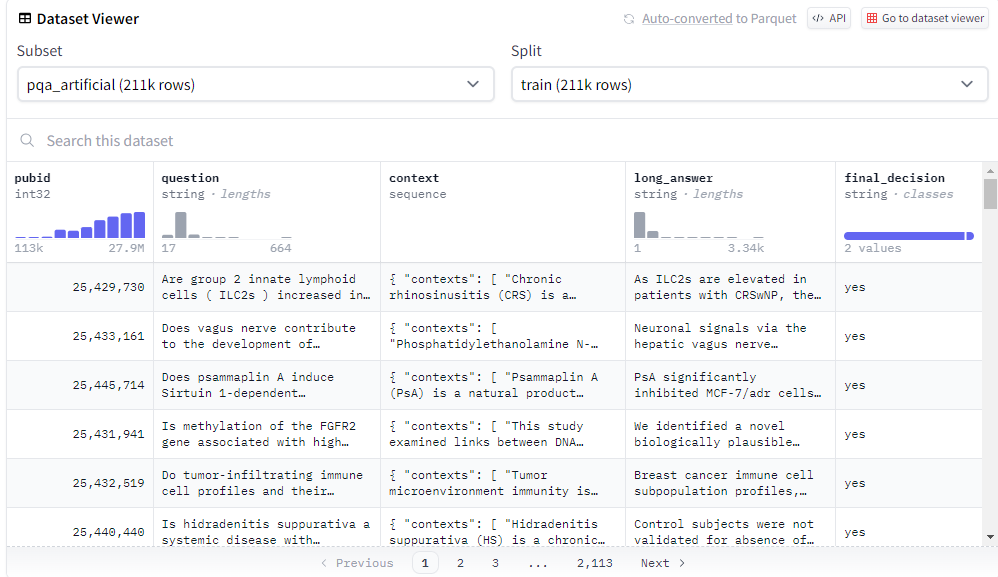
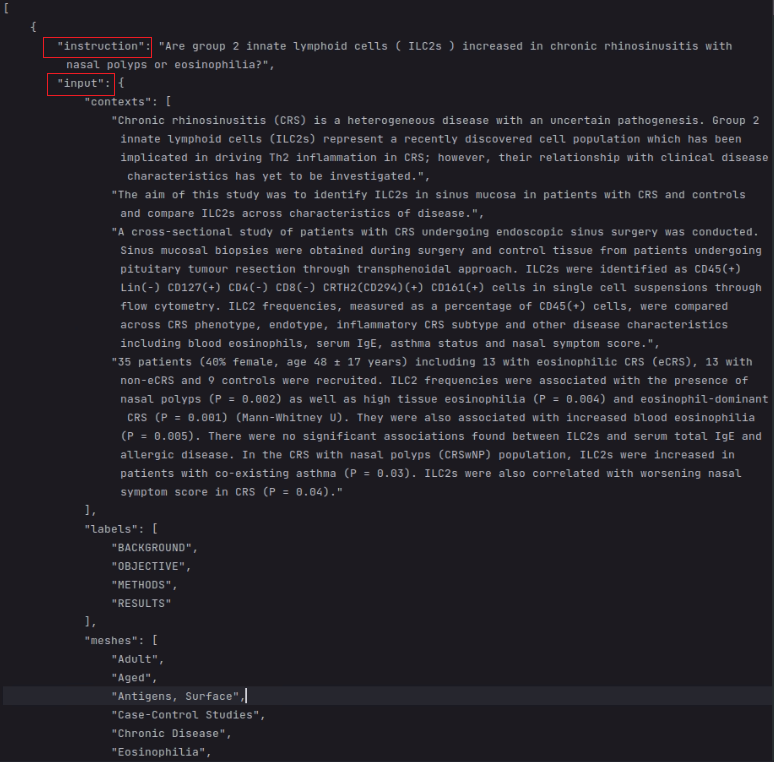
In this dataset, we set question as introduction, context as input, final_decion+long_answer as output.
click to view the code
from datasets import load_dataset
import os
import json
# Load the dataset
dataset = load_dataset("pubmed_qa", 'pqa_artificial')
# Define the save path
save_path = "../medical/PubMedQA" # Change this path to your local directory
os.makedirs(save_path, exist_ok=True)
# Function to save data as JSON with specified columns
def save_as_json(data, filename):
file_path = os.path.join(save_path, filename)
# Modify the data to include only 'question' and 'answer' columns
data_to_save = [{
"instruction": item['question'],
"input": str(item['context']),
"output": item['final_decision'] + ": " + item['long_answer']
} for item in data]
# Write the modified data to a JSON file
with open(file_path, 'w', encoding='utf-8') as f:
json.dump(data_to_save, f, ensure_ascii=False, indent=4)
# Save the modified data for train, validation, and test splits
save_as_json(dataset['train'], 'train.json')
# save_as_json(dataset['validation'], 'validation.json')
# save_as_json(dataset['test'], 'test.json')
3 fintuning commands
3.1 med_qa
click to view the code
CUDA_VISIBLE_DEVICES=1 python src/train_bash.py \
--stage sft \
--model_name_or_path ../llama/models_hf/7B \
--do_train \
--dataset alpaca_med_qa_en \
--template default \
--finetuning_type lora \
--lora_target q_proj,v_proj \
--output_dir ./FINE/llama2-7b-med_qa_single \
--overwrite_cache \
--per_device_train_batch_size 1 \
--gradient_accumulation_steps 4 \
--lr_scheduler_type cosine \
--logging_steps 10 \
--save_steps 1000 \
--learning_rate 5e-5 \
--num_train_epochs 3.0 \
--plot_loss \
--fp16
3.2 med_caq
click to view the code
CUDA_VISIBLE_DEVICES=2 python src/train_bash.py \
--stage sft \
--model_name_or_path ../llama/models_hf/7B \
--do_train \
--dataset alpaca_med_cqa_en \
--template default \
--finetuning_type lora \
--lora_target q_proj,v_proj \
--output_dir ./FINE/llama2-7b-med_cqa_single \
--overwrite_cache \
--per_device_train_batch_size 1 \
--gradient_accumulation_steps 4 \
--lr_scheduler_type cosine \
--logging_steps 10 \
--save_steps 1000 \
--learning_rate 5e-5 \
--num_train_epochs 3.0 \
--plot_loss \
--fp16
7B
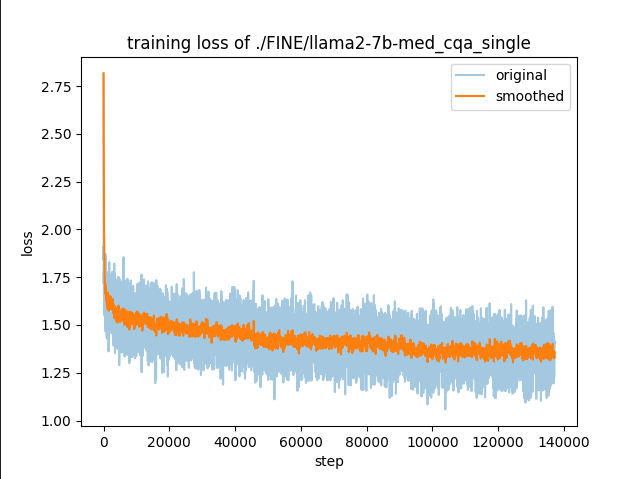
click to view the code
CUDA_VISIBLE_DEVICES=2 nohup python src/train_bash.py \
--stage sft \
--model_name_or_path ../llama/models_hf/13B \
--do_train \
--dataset alpaca_med_cqa_en \
--template default \
--finetuning_type lora \
--lora_target q_proj,v_proj \
--output_dir ./FINE/llama2-7b-med_cqa_single \
--overwrite_cache \
--per_device_train_batch_size 1 \
--gradient_accumulation_steps 4 \
--lr_scheduler_type cosine \
--logging_steps 10 \
--save_steps 1000 \
--learning_rate 5e-5 \
--num_train_epochs 3.0 \
--plot_loss \
--fp16 \
>> ./logs/fintuning_13B_medcaq_single.log 2>&1 &
3.3 pubmed_qa
click to view the code
CUDA_VISIBLE_DEVICES=3 python src/train_bash.py \
--stage sft \
--model_name_or_path ../llama/models_hf/7B \
--do_train \
--dataset alpaca_pubmed_qa_en \
--template default \
--finetuning_type lora \
--lora_target q_proj,v_proj \
--output_dir ./FINE/llama2-7b-pubmed_qa_single \
--overwrite_cache \
--per_device_train_batch_size 1 \
--gradient_accumulation_steps 4 \
--lr_scheduler_type cosine \
--logging_steps 10 \
--save_steps 1000 \
--learning_rate 5e-5 \
--num_train_epochs 3.0 \
--plot_loss \
--fp16
3.4 command for med_cqa
mixtral
click to view the code
accelerate launch src/train_bash.py \
--stage sft \
--do_train \
--model_name_or_path mistralai/Mixtral-8x7B-v0.1 \
--dataset alpaca_med_cqa_en \
--template default \
--quantization_bit 4 \
--lora_target q_proj,v_proj \
--output_dir ../FINE/mixtral-alpaca_med_cqa_en \
--overwrite_cache \
--per_device_train_batch_size 4 \
--gradient_accumulation_steps 4 \
--lr_scheduler_type cosine \
--logging_steps 10 \
--save_steps 1000 \
--learning_rate 5e-5 \
--num_train_epochs 3.0 \
--plot_loss


 浙公网安备 33010602011771号
浙公网安备 33010602011771号