数据分析-R语言学习笔记(三) 因子、缺失数据
一、因子
1、概念
首先,R中数据分为定量数据和定性数据:
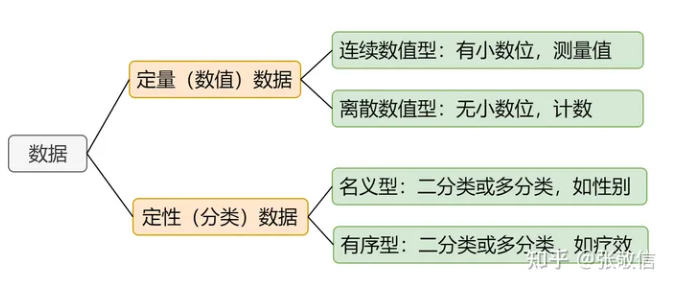
因子factor专门用来存放名义型和有序型的分类变量,因子本质上是一个带有水平level属性的整数向量,其中"水平"是指事前确定可能取值的有限集合,例如good,better,best。
一般数值型数据更容易是连续型数据,而字符串型数据更容易是名义型数据。
2、因子的应用
计算频数、独立性检验、相关性检验、方差分析、主成分分析、因子分析。
3、factor相关操作
对于因子,我们只需要知道数据集中哪些可以作为因子。这里拿mtcars数据集作为案例。
> mtcars$cyl [1] 6 6 4 6 8 6 8 4 4 6 6 8 8 8 8 8 8 4 4 4 4 8 8 8 8 4 4 4 8 6 8 4 > table(mtcars$cyl) #使用table()进行频数统计 4 6 8 11 7 14
使用factor()函数将向量转化为因子:
> f <- factor(c("red","red","green","red","blue"))
> f
[1] red red green red blue
Levels: blue green red
有序型变量作为因子:
> week <- factor(c("mon","tue","wed","thu","fri"))
> week
[1] mon tue wed thu fri
Levels: fri mon thu tue wed
> week <- factor(c("mon","tue","wed","thu","fri","sat","sun"))
> week
[1] mon tue wed thu fri sat sun
Levels: fri mon sat sun thu tue wed #输出没有顺序,而且可能是不全的
> week <- factor(c("mon","tue","wed","thu","fri","sat","sun"),ordered = T,levels = c("mon","tue","wed","thu","fri","sat","sun"))
> week
[1] mon tue wed thu fri sat sun
Levels: mon < tue < wed < thu < fri < sat < sun #此时输出有了顺序
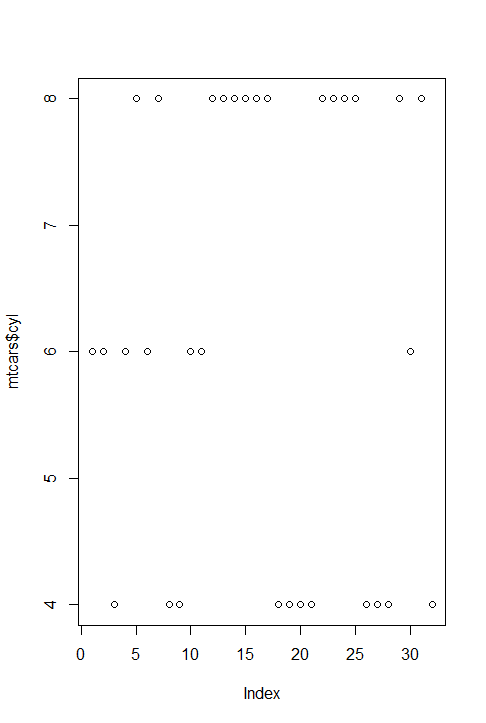
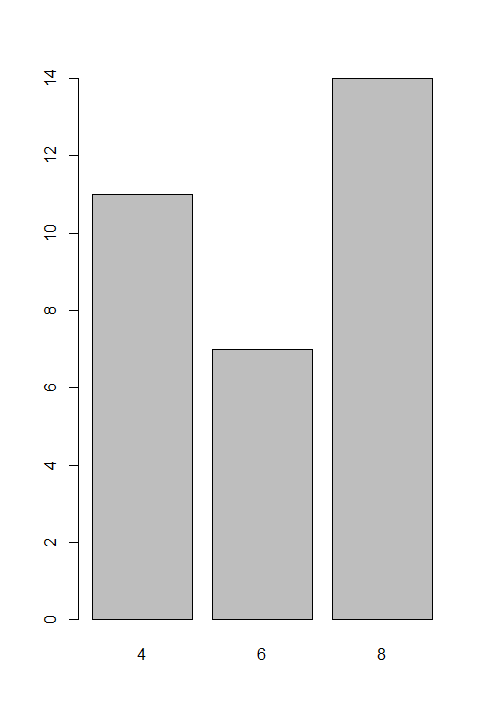
通过plot函数绘图,比较向量和因子的图像:
> plot(mtcars$cyl) #左图 > plot(factor(mtcars$cyl)) #右图
cut()函数:如果是个很大的数据集,我们就可以很容易通过cut计算每个区间包含的数据有多少,方便进行频数统计。
> num<-1:100 > num [1] 1 2 3 4 5 6 7 8 9 10 11 12 [13] 13 14 15 16 17 18 19 20 21 22 23 24 [25] 25 26 27 28 29 30 31 32 33 34 35 36 [37] 37 38 39 40 41 42 43 44 45 46 47 48 [49] 49 50 51 52 53 54 55 56 57 58 59 60 [61] 61 62 63 64 65 66 67 68 69 70 71 72 [73] 73 74 75 76 77 78 79 80 81 82 83 84 [85] 85 86 87 88 89 90 91 92 93 94 95 96 [97] 97 98 99 100 > cut(num,c(seq(0,100,10))) [1] (0,10] (0,10] (0,10] (0,10] (0,10] [6] (0,10] (0,10] (0,10] (0,10] (0,10] [11] (10,20] (10,20] (10,20] (10,20] (10,20] [16] (10,20] (10,20] (10,20] (10,20] (10,20] [21] (20,30] (20,30] (20,30] (20,30] (20,30] [26] (20,30] (20,30] (20,30] (20,30] (20,30] [31] (30,40] (30,40] (30,40] (30,40] (30,40] [36] (30,40] (30,40] (30,40] (30,40] (30,40] [41] (40,50] (40,50] (40,50] (40,50] (40,50] [46] (40,50] (40,50] (40,50] (40,50] (40,50] [51] (50,60] (50,60] (50,60] (50,60] (50,60] [56] (50,60] (50,60] (50,60] (50,60] (50,60] [61] (60,70] (60,70] (60,70] (60,70] (60,70] [66] (60,70] (60,70] (60,70] (60,70] (60,70] [71] (70,80] (70,80] (70,80] (70,80] (70,80] [76] (70,80] (70,80] (70,80] (70,80] (70,80] [81] (80,90] (80,90] (80,90] (80,90] (80,90] [86] (80,90] (80,90] (80,90] (80,90] (80,90] [91] (90,100] (90,100] (90,100] (90,100] (90,100] [96] (90,100] (90,100] (90,100] (90,100] (90,100] 10 Levels: (0,10] (10,20] (20,30] (30,40] ... (90,100]
state数据集中state.division和state.region是因子类型的数据。
二、缺失数据
出现缺失数据的原因:
机器断电,设备故障导致某个测量值发生了丢失;
测量根本没有发生,例如在做调查问卷时,有些问题没有回答,或者有些问题是无效的回答等等。
在R中,NA(not available),不可用,用来存储缺失信息。这里NA表示没有,但未必就一定是0,NA是不知道多少,可能是0可能不是。只要有缺失值出现的地方就返回NA值。
> 1+NA [1] NA > NA == 0 [1] NA
只要有缺失值出现的地方就返回NA值。这也会带来问题,只要数据集中有一个NA,整个数据集就不能用了:
> a <- c(NA,1:49) > a [1] NA 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 [27] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 > sum(a) [1] NA > mean(a) [1] NA
解决方法是,使用 na.rm 参数调整数据集:
> sum(a,na.rm =T) [1] 1225 > mean(a,na.rm=T) [1] 25
> mean(1:49,na.rm=T) #这说明将NA值移除后是按照49个向量计算的,移除NA,向量个数减少
[1] 25
R中提供is.na()进行逻辑测试:
> is.na(a) [1] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE [14] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE [27] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE [40] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
下面使用大一点的数据集做示例:
> library(VIM)
载入需要的程辑包:colorspace
载入需要的程辑包:grid
VIM is ready to use.
Suggestions and bug-reports can be submitted at: https://github.com/statistikat/VIM/issues
载入程辑包:‘VIM’
The following object is masked from ‘package:datasets’:
sleep
> sleep
BodyWgt BrainWgt NonD Dream Sleep Span Gest Pred Exp Danger
1 6654.000 5712.00 NA NA 3.3 38.6 645.0 3 5 3
2 1.000 6.60 6.3 2.0 8.3 4.5 42.0 3 1 3
3 3.385 44.50 NA NA 12.5 14.0 60.0 1 1 1
4 0.920 5.70 NA NA 16.5 NA 25.0 5 2 3
5 2547.000 4603.00 2.1 1.8 3.9 69.0 624.0 3 5 4
6 10.550 179.50 9.1 0.7 9.8 27.0 180.0 4 4 4
7 0.023 0.30 15.8 3.9 19.7 19.0 35.0 1 1 1
8 160.000 169.00 5.2 1.0 6.2 30.4 392.0 4 5 4
9 3.300 25.60 10.9 3.6 14.5 28.0 63.0 1 2 1
10 52.160 440.00 8.3 1.4 9.7 50.0 230.0 1 1 1
11 0.425 6.40 11.0 1.5 12.5 7.0 112.0 5 4 4
12 465.000 423.00 3.2 0.7 3.9 30.0 281.0 5 5 5
13 0.550 2.40 7.6 2.7 10.3 NA NA 2 1 2
14 187.100 419.00 NA NA 3.1 40.0 365.0 5 5 5
15 0.075 1.20 6.3 2.1 8.4 3.5 42.0 1 1 1
16 3.000 25.00 8.6 0.0 8.6 50.0 28.0 2 2 2
17 0.785 3.50 6.6 4.1 10.7 6.0 42.0 2 2 2
18 0.200 5.00 9.5 1.2 10.7 10.4 120.0 2 2 2
19 1.410 17.50 4.8 1.3 6.1 34.0 NA 1 2 1
20 60.000 81.00 12.0 6.1 18.1 7.0 NA 1 1 1
21 529.000 680.00 NA 0.3 NA 28.0 400.0 5 5 5
22 27.660 115.00 3.3 0.5 3.8 20.0 148.0 5 5 5
23 0.120 1.00 11.0 3.4 14.4 3.9 16.0 3 1 2
24 207.000 406.00 NA NA 12.0 39.3 252.0 1 4 1
25 85.000 325.00 4.7 1.5 6.2 41.0 310.0 1 3 1
26 36.330 119.50 NA NA 13.0 16.2 63.0 1 1 1
27 0.101 4.00 10.4 3.4 13.8 9.0 28.0 5 1 3
28 1.040 5.50 7.4 0.8 8.2 7.6 68.0 5 3 4
29 521.000 655.00 2.1 0.8 2.9 46.0 336.0 5 5 5
30 100.000 157.00 NA NA 10.8 22.4 100.0 1 1 1
31 35.000 56.00 NA NA NA 16.3 33.0 3 5 4
32 0.005 0.14 7.7 1.4 9.1 2.6 21.5 5 2 4
33 0.010 0.25 17.9 2.0 19.9 24.0 50.0 1 1 1
34 62.000 1320.00 6.1 1.9 8.0 100.0 267.0 1 1 1
35 0.122 3.00 8.2 2.4 10.6 NA 30.0 2 1 1
36 1.350 8.10 8.4 2.8 11.2 NA 45.0 3 1 3
37 0.023 0.40 11.9 1.3 13.2 3.2 19.0 4 1 3
38 0.048 0.33 10.8 2.0 12.8 2.0 30.0 4 1 3
39 1.700 6.30 13.8 5.6 19.4 5.0 12.0 2 1 1
40 3.500 10.80 14.3 3.1 17.4 6.5 120.0 2 1 1
41 250.000 490.00 NA 1.0 NA 23.6 440.0 5 5 5
42 0.480 15.50 15.2 1.8 17.0 12.0 140.0 2 2 2
43 10.000 115.00 10.0 0.9 10.9 20.2 170.0 4 4 4
44 1.620 11.40 11.9 1.8 13.7 13.0 17.0 2 1 2
45 192.000 180.00 6.5 1.9 8.4 27.0 115.0 4 4 4
46 2.500 12.10 7.5 0.9 8.4 18.0 31.0 5 5 5
47 4.288 39.20 NA NA 12.5 13.7 63.0 2 2 2
48 0.280 1.90 10.6 2.6 13.2 4.7 21.0 3 1 3
49 4.235 50.40 7.4 2.4 9.8 9.8 52.0 1 1 1
50 6.800 179.00 8.4 1.2 9.6 29.0 164.0 2 3 2
51 0.750 12.30 5.7 0.9 6.6 7.0 225.0 2 2 2
52 3.600 21.00 4.9 0.5 5.4 6.0 225.0 3 2 3
53 14.830 98.20 NA NA 2.6 17.0 150.0 5 5 5
54 55.500 175.00 3.2 0.6 3.8 20.0 151.0 5 5 5
55 1.400 12.50 NA NA 11.0 12.7 90.0 2 2 2
56 0.060 1.00 8.1 2.2 10.3 3.5 NA 3 1 2
57 0.900 2.60 11.0 2.3 13.3 4.5 60.0 2 1 2
58 2.000 12.30 4.9 0.5 5.4 7.5 200.0 3 1 3
59 0.104 2.50 13.2 2.6 15.8 2.3 46.0 3 2 2
60 4.190 58.00 9.7 0.6 10.3 24.0 210.0 4 3 4
61 3.500 3.90 12.8 6.6 19.4 3.0 14.0 2 1 1
62 4.050 17.00 NA NA NA 13.0 38.0 3 1 1
> is.na(sleep)
BodyWgt BrainWgt NonD Dream Sleep Span Gest Pred Exp Danger
[1,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[2,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[3,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[4,] FALSE FALSE TRUE TRUE FALSE TRUE FALSE FALSE FALSE FALSE
[5,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[6,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[7,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[8,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[9,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[10,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[11,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[12,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13,] FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE
[14,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[15,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[16,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[17,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[18,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[19,] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[20,] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[21,] FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE
[22,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[23,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[24,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[25,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[26,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[27,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[28,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[29,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[30,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[31,] FALSE FALSE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE
[32,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[33,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[34,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[35,] FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
[36,] FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE
[37,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[38,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[39,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[40,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[41,] FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE FALSE
[42,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[43,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[44,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[45,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[46,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[47,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[48,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[49,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[50,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[51,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[52,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[53,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[54,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[55,] FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[56,] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[57,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[58,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[59,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[60,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[61,] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[62,] FALSE FALSE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE
> colSums(sleep) # 使用colsums计算每一列缺失值数目
BodyWgt BrainWgt NonD Dream Sleep Span Gest Pred Exp Danger
12324.98 17554.32 NA NA NA NA NA 178.00 150.00 162.00
> rowSums(sleep)
[1] NA 77.700 NA NA 7862.800 428.650 96.723 776.800 152.900 794.560 163.825 1221.800 NA NA 66.575 129.200 79.685 163.000
[19] NA NA NA 333.260 55.820 NA 778.400 NA 77.701 110.540 1578.800 NA NA 53.445 117.060 1768.000 NA NA
[37] 57.023 65.978 67.800 179.600 NA 207.980 349.000 75.420 542.800 95.400 NA 61.280 139.035 405.000 264.250 274.400 NA 424.100
[55] NA NA 99.600 239.600 89.504 327.790 67.200 NA
na.omit()去掉数据集中的那些缺失值:
> c<-c(NA,1:20,NA,NA) > c [1] NA 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 NA NA > d<-na.omit(c) > d #最终结果没有NA值,还多了一些属性信息 [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 attr(,"na.action") [1] 1 22 23 attr(,"class") [1] "omit" > sum(d) [1] 210 > mean(d) [1] 10.5 > sum(c) [1] NA
na.omit()应用于数据框,则是将包含NA的一行都删掉:
> length(rownames(sleep)) [1] 62 > length(rownames(na.omit(sleep))) [1] 42
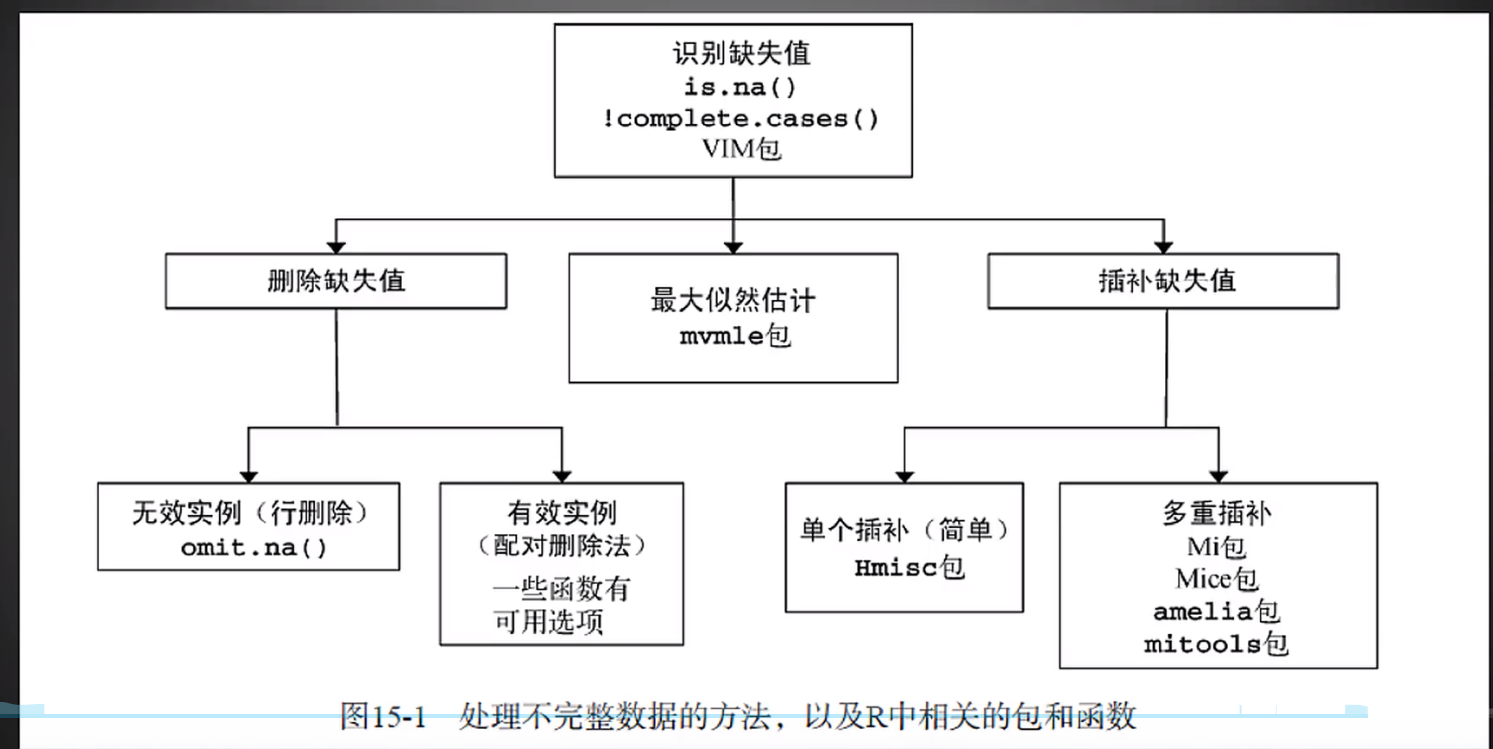
处理缺失值包:
其他缺失数据:
NaN,不可能值,不存在的;
Inf,无穷,分正负,存在的;
> -1/0 [1] -Inf > 0/0 [1] NaN > 1/0 [1] Inf > is.nan(0/0) [1] TRUE > is.-inf(-1/0) 错误: 找不到对象'is.' > is.infinite(-1/0) [1] TRUE



 浙公网安备 33010602011771号
浙公网安备 33010602011771号