plot a critical difference diagram , MATLAB code
建立criticaldifference函数
function cd = criticaldifference(s,labels,alpha)
%
% CRITICALDIFFERNCE - plot a critical difference diagram
%
% CRITICALDIFFERENCE(S,LABELS) produces a critical difference diagram [1]
% displaying the statistical significance (or otherwise) of a matrix of
% scores, S, achieved by a set of machine learning algorithms. Here
% LABELS is a cell array of strings giving the name of each algorithm.
%
% References
%
% [1] Demsar, J., "Statistical comparisons of classifiers over multiple
% datasets", Journal of Machine Learning Research, vol. 7, pp. 1-30,
% 2006.
%
%
% File : criticaldifference.m
%
% Date : Monday 14th April 2008
%
% Author : Gavin C. Cawley
%
% Description : Sparse multinomial logistic regression using a Laplace prior.
%
% History : 14/04/2008 - v1.00
%
% Copyright : (c) Dr Gavin C. Cawley, April 2008.
%
% This program is free software; you can redistribute it and/or modify
% it under the terms of the GNU General Public License as published by
% the Free Software Foundation; either version 2 of the License, or
% (at your option) any later version.
%
% This program is distributed in the hope that it will be useful,
% but WITHOUT ANY WARRANTY; without even the implied warranty of
% MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
% GNU General Public License for more details.
%
% You should have received a copy of the GNU General Public License
% along with this program; if not, write to the Free Software
% Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
%
% Thanks to Gideon Dror for supplying the extended table of critical values.
if nargin < 3
alpha = 0.1;
end
% convert scores into ranks
[N,k] = size(s);
[S,r] = sort(s');
idx = k*repmat(0:N-1, k, 1)' + r';
R = repmat(1:k, N, 1);
S = S';
for i=1:N
for j=1:k
index = S(i,j) == S(i,:);
R(i,index) = mean(R(i,index));
end
end
r(idx) = R;
r = r';
% compute critical difference
if alpha == 0.01
qalpha = [0.000 2.576 2.913 3.113 3.255 3.364 3.452 3.526 3.590 3.646 ...
3.696 3.741 3.781 3.818 3.853 3.884 3.914 3.941 3.967 3.992 ...
4.015 4.037 4.057 4.077 4.096 4.114 4.132 4.148 4.164 4.179 ...
4.194 4.208 4.222 4.236 4.249 4.261 4.273 4.285 4.296 4.307 ...
4.318 4.329 4.339 4.349 4.359 4.368 4.378 4.387 4.395 4.404 ...
4.412 4.420 4.428 4.435 4.442 4.449 4.456 ];
elseif alpha == 0.05
qalpha = [0.000 1.960 2.344 2.569 2.728 2.850 2.948 3.031 3.102 3.164 ...
3.219 3.268 3.313 3.354 3.391 3.426 3.458 3.489 3.517 3.544 ...
3.569 3.593 3.616 3.637 3.658 3.678 3.696 3.714 3.732 3.749 ...
3.765 3.780 3.795 3.810 3.824 3.837 3.850 3.863 3.876 3.888 ...
3.899 3.911 3.922 3.933 3.943 3.954 3.964 3.973 3.983 3.992 ...
4.001 4.009 4.017 4.025 4.032 4.040 4.046];
elseif alpha == 0.1
qalpha = [0.000 1.645 2.052 2.291 2.460 2.589 2.693 2.780 2.855 2.920 ...
2.978 3.030 3.077 3.120 3.159 3.196 3.230 3.261 3.291 3.319 ...
3.346 3.371 3.394 3.417 3.439 3.459 3.479 3.498 3.516 3.533 ...
3.550 3.567 3.582 3.597 3.612 3.626 3.640 3.653 3.666 3.679 ...
3.691 3.703 3.714 3.726 3.737 3.747 3.758 3.768 3.778 3.788 ...
3.797 3.806 3.814 3.823 3.831 3.838 3.846];
else
error('alpha must be 0.01, 0.05 or 0.1');
end
cd = qalpha(k)*sqrt(k*(k+1)/(6*N));
figure(1);
clf
axis off
axis([-0.2 1.2 -20 140]);
axis xy
tics = repmat((0:(k-1))/(k-1), 3, 1);
line(tics(:), repmat([100, 101, 100], 1, k), 'LineWidth', 1.5, 'Color', 'k');
%tics = repmat(((0:(k-2))/(k-1)) + 0.5/(k-1), 3, 1);
%line(tics(:), repmat([100, 101, 100], 1, k-1), 'LineWidth', 1.5, 'Color', 'k');
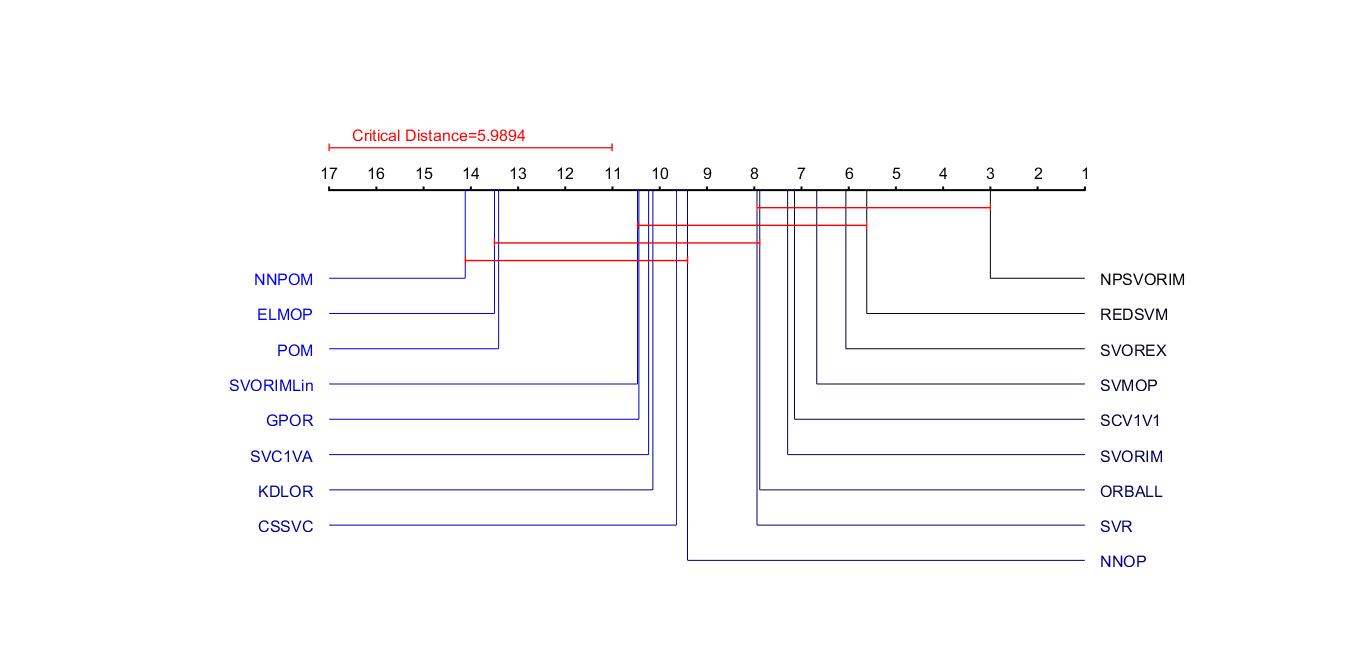
line([0 0 0 cd/(k-1) cd/(k-1) cd/(k-1)], [113 111 112 112 111 113], 'LineWidth', 1, 'Color', 'r');
text(0.03, 116, ['Critical Distance=' num2str(cd)], 'FontSize', 12, 'HorizontalAlignment', 'left', 'Color', 'r');
for i=1:k
text((i-1)/(k-1), 105, num2str(k-i+1), 'FontSize', 12, 'HorizontalAlignment', 'center');
end
% compute average ranks
r = mean(r);
[r,idx] = sort(r);
% compute statistically similar cliques
clique = repmat(r,k,1) - repmat(r',1,k);
clique(clique<0) = realmax;
clique = clique < cd;
for i=k:-1:2
if all(clique(i-1,clique(i,:))==clique(i,clique(i,:)))
clique(i,:) = 0;
end
end
n = sum(clique,2);
clique = clique(n>1,:);
n = size(clique,1);
%yanse={'b','g','y','m','r'};
b=linspace(0,1,k);
% labels displayed on the right
for i=1:ceil(k/2)
line([(k-r(i))/(k-1) (k-r(i))/(k-1) 1], [100 100-3*(n+1)-10*i 100-3*(n+1)-10*i], 'Color', [0 0 b(i)]);
%text(1.2, 100 - 5*(n+1)- 10*i + 2, num2str(r(i)), 'FontSize', 10, 'HorizontalAlignment', 'right');
text(1.02, 100 - 3*(n+1) - 10*i, labels{idx(i)}, 'FontSize', 12, 'VerticalAlignment', 'middle', 'HorizontalAlignment', 'left', 'Color', [0 0 b(i)]);
end
% labels displayed on the left
for i=ceil(k/2)+1:k
line([(k-r(i))/(k-1) (k-r(i))/(k-1) 0], [100 100-3*(n+1)-10*(k-i+1) 100-3*(n+1)-10*(k-i+1)], 'Color', [0 0 b(i)]);
%text(-0.2, 100 - 5*(n+1) -10*(k-i+1)+2, num2str(r(i)), 'FontSize', 10, 'HorizontalAlignment', 'left');
text(-0.02, 100 - 3*(n+1) -10*(k-i+1), labels{idx(i)}, 'FontSize', 12, 'VerticalAlignment', 'middle', 'HorizontalAlignment', 'right', 'Color', [0 0 b(i)]);
end
% group cliques of statistically similar classifiers
for i=1:size(clique,1)
R = r(clique(i,:));
%line([((k-min(R))/(k-1)) + 0.015 ((k - max(R))/(k-1)) - 0.015], [100-5*i 100-5*i], 'LineWidth', 1, 'Color', 'r');
%line([0 0 0 cd/(k-1) cd/(k-1) cd/(k-1)], [113 111 112 112 111 113], 'LineWidth', 1, 'Color', 'r');
line([((k-min(R))/(k-1)) ((k-min(R))/(k-1)) ((k-min(R))/(k-1)) ((k - max(R))/(k-1)) ((k - max(R))/(k-1)) ((k - max(R))/(k-1))], [100+1-5*i 100-1-5*i 100-5*i 100-5*i 100-1-5*i 100+1-5*i], 'LineWidth', 1, 'Color', 'r');
end
可执行m文件:
load Data
s=AccMatrix;
labels={'SCV1V1','SVC1VA','SVR','CSSVC','SVMOP','NNOP','ELMOP','POM',...
'NNPOM', 'SVOREX','SVORIM','SVORIMLin','KDLOR','GPOR','REDSVM','ORBALL' ,'NPSVORIM'};%方法的标签
alpha=0.05; %显著性水平0.1,0.05或0.01
cd = criticaldifference(s,labels,alpha)
AccMatrix=[
0.28 0.12 0.28 0.11 0.32 0.08 0.26 0.13 0.37 0.10 0.28 0.12 0.42 0.21 0.38 0.17 0.36 0.14 0.36 0.13 0.38 0.12 0.37 0.10 0.34 0.15 0.39 0.09 0.37 0.12 0.36 0.13 0.37 0.11
0.31 0.12 0.33 0.11 0.34 0.13 0.32 0.11 0.32 0.09 0.24 0.11 0.40 0.18 0.50 0.15 0.34 0.18 0.35 0.12 0.34 0.12 0.34 0.12 0.33 0.11 0.48 0.17 0.33 0.11 0.30 0.12 0.28 0.14
0.36 0.09 0.40 0.14 0.39 0.11 0.39 0.13 0.40 0.09 0.39 0.11 0.44 0.16 0.62 0.15 0.50 0.13 0.37 0.13 0.37 0.13 0.37 0.13 0.39 0.12 0.55 0.10 0.38 0.13 0.36 0.12 0.32 0.10
0.22 0.12 0.28 0.16 0.24 0.10 0.27 0.15 0.27 0.11 0.29 0.11 0.39 0.13 0.65 0.14 0.39 0.14 0.26 0.11 0.27 0.11 0.32 0.11 0.26 0.11 0.36 0.16 0.27 0.12 0.30 0.10 0.22 0.10
0.44 0.06 0.45 0.06 0.40 0.07 0.43 0.07 0.46 0.06 0.41 0.06 0.44 0.08 0.50 0.08 0.45 0.09 0.41 0.07 0.40 0.07 0.48 0.07 0.43 0.05 0.67 0.04 0.40 0.07 0.40 0.06 0.41 0.05
0.03 0.03 0.04 0.03 0.04 0.02 0.04 0.02 0.04 0.03 0.04 0.02 0.06 0.02 0.03 0.02 0.03 0.03 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.04 0.03 0.03 0.03
0.03 0.01 0.03 0.01 0.16 0.03 0.03 0.01 0.03 0.01 0.04 0.01 0.09 0.02 0.09 0.02 0.06 0.05 0.00 0.01 0.00 0.01 0.09 0.02 0.16 0.03 0.03 0.01 0.00 0.00 0.03 0.02 0.02 0.01
0.42 0.03 0.44 0.03 0.43 0.03 0.43 0.03 0.42 0.03 0.42 0.03 0.43 0.02 0.43 0.03 0.46 0.03 0.43 0.03 0.43 0.03 0.43 0.03 0.51 0.03 0.42 0.03 0.43 0.03 0.44 0.03 0.43 0.03
0.01 0.00 0.01 0.01 0.03 0.01 0.01 0.01 0.00 0.00 0.03 0.01 0.16 0.01 0.84 0.30 0.11 0.02 0.01 0.01 0.01 0.01 0.08 0.01 0.05 0.01 0.04 0.01 0.01 0.00 0.01 0.01 0.01 0.00
0.43 0.04 0.43 0.06 0.46 0.07 0.43 0.06 0.45 0.10 0.53 0.09 0.57 0.13 0.66 0.16 0.62 0.14 0.45 0.06 0.45 0.07 0.43 0.08 0.47 0.09 0.42 0.03 0.44 0.05 0.46 0.09 0.42 0.08
0.05 0.03 0.05 0.03 0.07 0.04 0.05 0.03 0.07 0.03 0.06 0.03 0.07 0.03 0.71 0.03 0.06 0.03 0.02 0.01 0.02 0.01 0.74 0.01 0.11 0.03 0.05 0.02 0.02 0.01 0.05 0.02 0.04 0.03
0.36 0.03 0.45 0.03 0.36 0.03 0.44 0.03 0.35 0.03 0.42 0.04 0.43 0.03 0.85 0.02 0.46 0.04 0.36 0.03 0.36 0.03 0.36 0.02 0.37 0.03 0.31 0.03 0.36 0.03 0.38 0.03 0.34 0.03
0.37 0.02 0.37 0.02 0.38 0.02 0.37 0.02 0.37 0.02 0.37 0.03 0.37 0.02 0.38 0.03 0.38 0.02 0.38 0.02 0.38 0.02 0.39 0.02 0.46 0.03 0.39 0.03 0.37 0.02 0.39 0.03 0.37 0.03
0.25 0.06 0.26 0.06 0.32 0.07 0.27 0.06 0.26 0.04 0.39 0.06 0.38 0.06 0.53 0.19 0.55 0.08 0.32 0.05 0.32 0.07 0.41 0.07 0.30 0.07 0.39 0.07 0.32 0.07 0.29 0.05 0.27 0.05
0.35 0.02 0.36 0.02 0.37 0.02 0.36 0.02 0.36 0.02 0.40 0.02 0.40 0.02 0.40 0.02 0.40 0.02 0.37 0.02 0.37 0.02 0.41 0.02 0.35 0.02 0.39 0.01 0.37 0.02 0.33 0.02 0.36 0.02
0.31 0.04 0.33 0.03 0.30 0.03 0.32 0.03 0.29 0.03 0.31 0.04 0.30 0.04 0.29 0.03 0.34 0.13 0.29 0.03 0.28 0.03 0.29 0.04 0.36 0.03 0.29 0.03 0.29 0.03 0.32 0.02 0.29 0.03
0.74 0.02 0.82 0.03 0.75 0.02 0.80 0.03 0.74 0.02 0.71 0.02 0.75 0.02 0.74 0.02 0.73 0.03 0.71 0.03 0.75 0.02 0.76 0.02 0.81 0.03 0.71 0.03 0.75 0.02 0.76 0.02 0.75 0.03 ];



 浙公网安备 33010602011771号
浙公网安备 33010602011771号