![]()
# 参考1:https://github.com/digitalcytometry/cytotrace2?tab=readme-ov-file
# 参考2:http://www.360doc.com/content/24/0426/09/65403234_1121488444.shtml
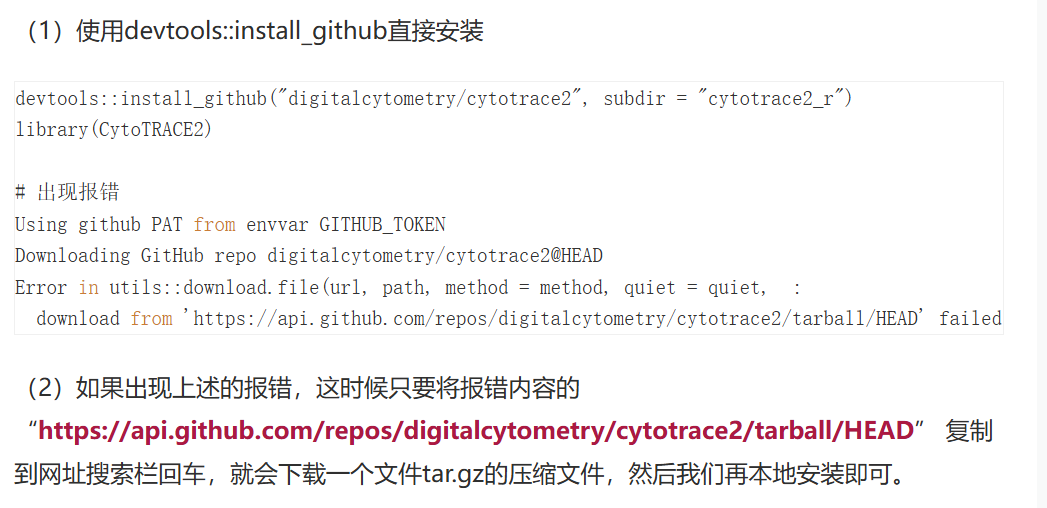
devtools::install_github("digitalcytometry/cytotrace2", subdir = "cytotrace2_r") #installing #失败
#本地安装
remotes::install_local("./Downloads/digitalcytometry-cytotrace2-v1.1.0-10-g23eaec2.tar.gz",
subdir="cytotrace2_r",#要具体查看
upgrade =F,dependencies=T)
# tar -tzf ./Downloads/digitalcytometry-cytotrace2-v1.1.0-10-g23eaec2.tar.gz
# 查看对应的subdir命名
library(CytoTRACE2)
library(tidyverse)
library(Seurat)
setwd("./cytotrace/test")
# load the CytoTRACE 2 package
library(CytoTRACE2)
# download the .rds file
# download.file("https://drive.google.com/uc?export=download&id=1TYdQsMoDIJjoeuiTD5EO_kZgNJUyfRY2", "Pancreas_10x_downsampled.rds")
# 手动下载
# load rds
data <- readRDS("Pancreas_10x_downsampled.rds")
# extract expression data
expression_data <- data$expression_data
# running CytoTRACE 2 main function - cytotrace2 - with default parameters
cytotrace2_result <- cytotrace2(expression_data)
# extract annotation data
annotation <- data$annotation
# generate prediction and phenotype association plots with plotData function
plots <- plotData(cytotrace2_result = cytotrace2_result,
annotation = annotation,
expression_data = expression_data)
# 其余图略



 浙公网安备 33010602011771号
浙公网安备 33010602011771号