微生物分类学与命名法
https://en.wikipedia.org/wiki/Bacterial_taxonomy
https://www.dsmz.de/services/online-tools/prokaryotic-nomenclature-up-to-date
nomenclature 命名法
Taxonomy 分类学
LPSN - List of Prokaryotic names with Standing in Nomenclature
NCBI taxonomy
Bergey’s
The List of Prokaryotic names with Standing in Nomenclature (LPSN) provides comprehensive information on the nomenclature of prokaryotes and much more. Navigating LPSN is easy.LPSN is a free to use service founded by Jean P. Euzéby in 1997 and later on maintained by Aidan C. Parte.As of 2020, the regularly augmented Prokaryotic Nomenclature Up-to-date (PNU) database will form the basis of a new LPSN-PNU merged service. PNU was established in 1993 as a service of the Leibniz Institute DSMZ, and was curated by Norbert Weiss, Manfred Kracht and Dorothea Gleim.
How to cite LPSN
- Parte, A.C. (2018). LPSN — List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. International Journal of Systematic and Evolutionary Microbiology, 68, 1825-1829; doi: 10.1099/ijsem.0.002786
- Parte, A.C. (2013). LPSN — List of Prokaryotic names with Standing in Nomenclature. Nucleic Acids Research, 42, Issue D1, D613–D616; doi: 10.1093/nar/gkt1111
- Euzéby, J.P. (1997). List of Bacterial Names with Standing in Nomenclature: a Folder Available on the Internet. International Journal of Systematic Bacteriology, 47, 590-592; doi:10.1099/00207713-47-2-590
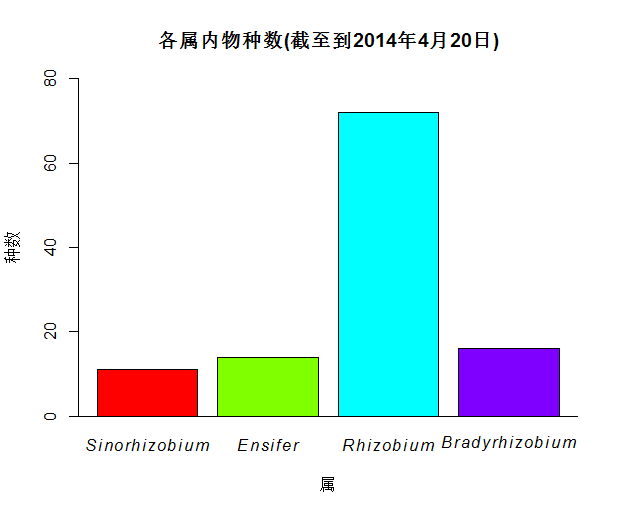
用R语言统计一下LPSN网站上的各属内的物种数
#使用R语言,可以将LPSN网站(http://www.bacterio.net)上的感兴趣的细菌的属下的物种数统计出来并做图:
library(stringr)
#统计中华根瘤菌属(Sinorhizobium)
SinorhizobiumPage <- readLines("http://www.bacterio.net/sinorhizobium.html")
grep('Number of species', SinorhizobiumPage)
pat1 <- "(\d)+1"
(SinorhizobiumSpNum <- as.numeric(str_extract(SinorhizobiumPage[125], pat1)))
#统计剑菌属(Ensifer)
EnsiferPage <- readLines("http://www.bacterio.net/ensifer.html")
grep('Number of species', EnsiferPage)
EnsiferPage[125]
pat2 <- "(\d)+4"
(EnsiferSpNum <- as.numeric(str_extract(EnsiferPage[125], pat2)))
#统计根瘤菌属(Rhizobium)
RhizobiumPage <- readLines("http://www.bacterio.net/rhizobium.html")
grep('Number of species', RhizobiumPage)
pat3 <- "(\d)+2"
(RhizobiumSpNum <- as.numeric(str_extract(RhizobiumPage[125], pat3)))
#统计慢生根瘤菌属(Bradyrhizobium)
BradyrhizobiumPage <- readLines("http://www.bacterio.net/bradyrhizobium.html")
grep('Number of species', BradyrhizobiumPage)
pat4 <- "(\d)+6"
(BradyrhizobiumSpNum <- as.numeric(str_extract(BradyrhizobiumPage[125], pat4)))
rhizobiaNumbers <- data.frame(S=SinorhizobiumSpNum, E=EnsiferSpNum, R=RhizobiumSpNum,
B=BradyrhizobiumSpNum)
sino <- expression(italic(Sinorhizobium))
ensi <- expression(italic(Ensifer))
rhi <- expression(italic(Rhizobium))
brady <- expression(italic(Bradyrhizobium))
t.rhizobia <- t(rhizobiaNumbers)
t1.rhizobia <- as.data.frame(t.rhizobia)
barp <- barplot(t1.rhizobia$V1, names.arg=c(sino, ensi, rhi, brady),
xlab="属", ylab="种数", col=rainbow(4),
main="各属内根瘤菌的种数(截至到2014年4月20日)",
ylim=c(0, 80))
abline(h=0)
#经做图后如下:



 浙公网安备 33010602011771号
浙公网安备 33010602011771号