现在想尝试一下,如果我们将人特异性的序列分开来看,是否可以得到一些不太一样的信息?
现在先用SVA_D做一个测试。
1。提取SVA_D的人特异性的序列。
grep 'SVA_D' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s+/\t/g' |bgzip -c >Hs_SVA_D.bed.gz
想看看能不能找到一种在人脑中特异性富集的人特异性插入的重复序列的家族,如果能够找到,则是一个非常有用的信息。
2。比对和画图
time giggle search -i split_sort_b -q Hs_SVA_D.bed.gz -s >Hs_SVA_D.bed.gz.giggle.result
real 0m0.214s
user 0m0.106s
sys 0m0.081s
python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_D.bed.gz.giggle.result -o Hs_SVA_D.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
结果为空,反思应该是输入文件格式的问题。因为从道理上讲,我们的reference是包括一些静息的位置的,因此,不可能全部都是为0的情况。
现在重新反思调整。
调整了一下这行代码。
grep 'SVA_D' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_D.bed.gz
和全部的SVA比起来,感觉特征非常的相似。
什么家族的divergence是比较大的呢?
SVA-A
(base) [xxzhang@mu02 Roadmap]$ grep 'SVA_A' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_A.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_SVA_A.bed.gz -s >Hs_SVA_A.bed.gz.giggle.result
real 0m0.102s
user 0m0.018s
sys 0m0.044s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_A.bed.gz.giggle.result -o Hs_SVA_A.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-2.7961962495046206 90.29874405790137
SVA-B
(base) [xxzhang@mu02 Roadmap]$ grep 'SVA_B' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_B.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_SVA_B.bed.gz -s >Hs_SVA_B.bed.gz.giggle.result
real 0m0.080s
user 0m0.022s
sys 0m0.022s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_B.bed.gz.giggle.result -o Hs_SVA_B.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-2.802828875395419 122.58224670288607
SVA-C
(base) [xxzhang@mu02 Roadmap]$ grep 'SVA_C' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_C.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_SVA_C.bed.gz -s >Hs_SVA_C.bed.gz.giggle.result
real 0m0.118s
user 0m0.038s
sys 0m0.027s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_C.bed.gz.giggle.result -o Hs_SVA_C.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-11.346883898618795 379.3057133827626
SVA-E
(base) [xxzhang@mu02 Roadmap]$ grep 'SVA_E' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_E.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_SVA_E.bed.gz -s >Hs_SVA_E.bed.gz.giggle.result
real 0m0.222s
user 0m0.060s
sys 0m0.087s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_E.bed.gz.giggle.result -o Hs_SVA_E.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-44.21506626398062 77.89403691192818
SVA-F
(base) [xxzhang@mu02 Roadmap]$ grep 'SVA_F' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_SVA_F.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_SVA_F.bed.gz -s >Hs_SVA_F.bed.gz.giggle.result
real 0m0.368s
user 0m0.156s
sys 0m0.079s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_SVA_F.bed.gz.giggle.result -o Hs_SVA_F.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-118.86515607858043 441.02930560324324
接下来就继续用不同的小的家族的序列来做测试。
AluYa5
(base) [xxzhang@mu02 Roadmap]$ grep 'AluYa5' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_AluYa5.bed.gz (base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_AluYa5.bed.gz -s >Hs_AluYa5.bed.gz.giggle.result
real 0m1.331s
user 0m0.683s
sys 0m0.257s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_AluYa5.bed.gz.giggle.result -o Hs_AluYa5.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-159.19499252187038 120.78549515881221
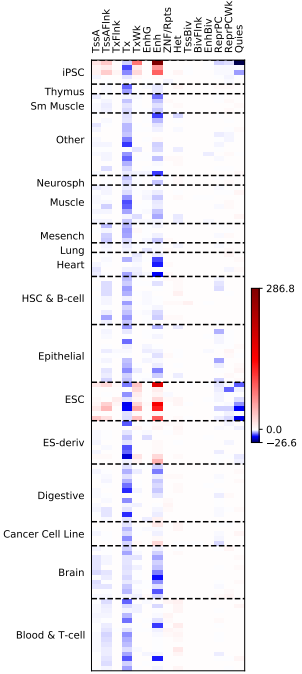
LTR5Hs
(base) [xxzhang@mu02 Roadmap]$ grep 'LTR5_Hs' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's/\s\+/\t/g' |bgzip -c >Hs_LTR5Hs.bed.gz
(base) [xxzhang@mu02 Roadmap]$ time giggle search -i split_sort_b -q Hs_LTR5Hs.bed.gz -s >Hs_LTR5Hs.bed.gz.giggle.result
real 0m0.234s
user 0m0.061s
sys 0m0.099s
(base) [xxzhang@mu02 Roadmap]$ python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_LTR5Hs.bed.gz.giggle.result -o Hs_LTR5Hs.bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt
-26.615647364611128 286.7638692827507
在LTR5Hs这里,似乎得到了一个比较有意思的结果(并且是人家已经发现过的)。
我现在考虑用代码批量的传入参数,看看批量运行,都看一看有哪些的pattern。(我想找到那个在人脑中特异性表达的那个重复序列,亲爱的,我想把你找到)
提到这个,我好像之前做过这样的尝试,我当时是使用了perl的一个package来实现的。
现在也重新用这个思路来做。
awk '{print $4}' /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sort |uniq >Hs_repeat_subfamily.txt
刚刚借鉴了之前的代码,使用perl的system()函数,批量的嵌套shell的指令,进行循环,还挺好用的。
#!perl
open (MARK, "< Hs_repeat_subfamily.txt") or die "can not open it!";
while ($line = <MARK>){
print($line);
chomp($line);
print($line);
system_call("grep \"".$line."\" /home/xxzhang/workplace/project/geneRegion/repeat_interest.bed |sed 's\/\\s\\+\/\\t\/g' |bgzip -c >Hs_".$line.".bed.gz");
system_call("time giggle search -i split_sort_b -q Hs_".$line.".bed.gz -s >Hs_".$line.".bed.gz.giggle.result");
system_call("python /home/xxzhang/workplace/software/giggle/scripts/giggle_heat_map.py -s /home/xxzhang/workplace/software/giggle/examples/rme/states.txt -c /home/xxzhang/workplace/software/giggle/examples/rme/EDACC_NAME.txt -i Hs_".$line.".bed.gz.giggle.result -o ./result/Hs_".$line.".bed.gz.3x11.pdf -n /home/xxzhang/workplace/software/giggle/examples/rme/new_groups.txt --x_size 3 --y_size 11 --stat combo --ablines 15,26,31,43,52,60,72,82,87,89,93,101,103,116,120,122,127 --state_names /home/xxzhang/workplace/software/giggle/examples/rme/short_states.txt --group_names /home/xxzhang/workplace/software/giggle/examples/rme/new_groups_names.txt");
}
close(MARK);
sub system_call
{
my $command=$_[0];
print "\n\n".$command."\n\n";
system($command);
}
然后qsub放到服务器上运行,6分钟左右就把所有的家族的结果弄出来了。这部分的结果,我明天来看。
刚刚粗粗看了一下,还是找到一些挺有意思的pattern的。
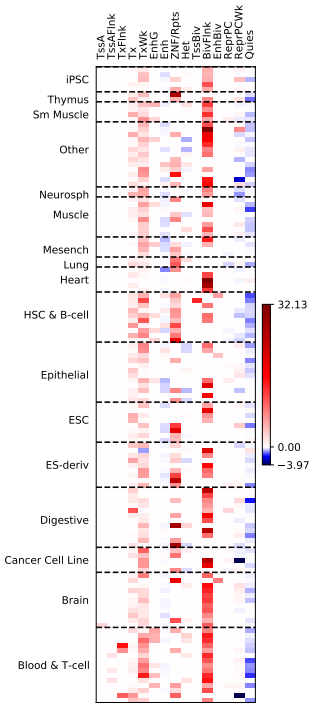
今天的任务结束。就是这样一点点的探索来的。
相信一定有意义,只是自己还没有探索出来,不要怕,一点点的往前走。
明天使用cistrome的数据试一下。




 浙公网安备 33010602011771号
浙公网安备 33010602011771号