GA算法-R语言实现
GA算法-R语言实现
旅行商问题
班共有30位同学,来自22个地区,我们希望在假期来一次说走就走的旅行,将所有同学的家乡走一遍。算起来,路费是一笔很大的花销,所以希望设计一个旅行方案,确保这一趟走下来的总路程最短。
旅行商问题是一个经典的NP问题
NP就是Non-deterministic Polynomial,即多项式复杂程度的非确定性问题,是世界七大数学难题之一。
如果使用枚举法求解,22个地点共有:
(22-1)!/2 = 25545471085854720000 种路线方案
GA算法
遗传算法将“优胜劣汰,适者生存”的生物进化原理引入优化参数形成的编码串联群体中,按所选择的适应度函数并通过遗传中的复制、交叉及变异对个体进行筛选,使适应度高的个体被保留下来,组成新的群体,新的群体既继承了上一代的信息,又优于上一代。这样周而复始,群体中个体适应度不断提高,直到满足一定的条件。遗传算法的算法简单,可并行处理,并能到全局最优解。
GA算法设计
1.生成原始染色体种群
采用实数编码,以N个城市的序号作为一条可能的路径。 例如对8个城市,可生成如下的染色体代表一条路径,8,6,4,2,7,5,3,1.重复操作生成数目等于n的染色体种群。
2.生成适应度函数
由于是求最短路径,适应度函数一般求函数最大值,所以取路径总长度T的倒数,即fitness=1/T。
3.选择染色体
采用轮盘赌的方式产生父代染色体。
4.对染色体种群进行编码
假设有一个含有九个城市的列表:W=(A,B,C,D,E,F,G,H,I)。
有如下两条路线:
W1=(A,D,B,H,F,I,G,E,C)
W2=(B,C,A,D,E,H,I,F,G)
则这两条路线可编码为:
W1=(142869753)
W2=(231458967)
5.交叉
以概率Pc选择参加交叉的个体(偶数个),用两点交叉算子进行操作。
例如对于下面两个染色体个体
(1 3 4 | 5 2 9 | 8 6 7)
(1 7 6 | 9 5 2 | 4 3 8)
通过两点交叉可得到子代染色体为
(1 3 4 | 9 5 2 | 8 6 7)
(1 7 6 | 5 2 9 | 4 3 8)
6.变异
以概率Pm选择参加变异的个体,用对换变异进行操作。随机的选择个体中的两个位点,进行交换基因。
如A=123456789;如果对换点为4和7,则经过对换后为B=123756489
7.解码
对染色体进行解码,恢复染色体的实数表示方法。
8.逐代进化
根据得出的新的染色体,再次返回选择染色体的步骤,进行迭代,直到达到迭代次数,算法停止。
算法实现
#加载packages
library(sp)
library(maptools)
library(geosphere)
source("C:\\Users\\ShangFR\\Desktop\\路径优化\\GA算法脚本.R")
data=read.csv("C:\\Users\\ShangFR\\Desktop\\路径优化\\143地理坐标.csv") #读取城市经纬度数据
border <- readShapePoly("C:\\Users\\ShangFR\\Desktop\\路径优化\\map\\bou2_4p.shp") #读取各省的边界数据等
#初始化(列出地区距离矩阵-聚类)
da=data[,1:2]
rownames(da)=data[,3]
hc=hclust(dist(da))
cutree(hc, h = 10)
plot(hc)
route=CreatDNA(data,5)
x = route[,1]
y = route[,2]
z = route[,3]
cols=route[,4]
muer.lonlat = cbind(route[,1],route[,2]) # matrix
muer.dists = distm(muer.lonlat, fun=distVincentyEllipsoid) # 精确计算,椭圆
ans=round(muer.dists/1000,2)
roundots = list(x=x,y=y,ans=ans,z=z,cols=cols)
species = GA4TSP(dots=roundots,initDNA=NULL,N=50,cp=0.1,vp=0.01,maxIter=1000,maxStay=100,maxElite=2,drawing=TRUE)
![]()
#粗糙地计算总路程上界UpperBound = function(disM){ mx = apply(disM,1,max) #两城市间最长距离 return( sum(mx) ) #最长旅行路径}#生成随机染色体rndDNA = function(n){ return( seq(1,n,1) ) #随机生成n条旅行路径}#对一个解计算总路程的距离calcScores = function(dna,disM){ n = length(dna) #城市总数 tmp = cbind(dna[1:n],c(dna[2:n],dna[1])) #生成起始点城市 len = apply(tmp,1,function(x) disM[x[1],x[2]]) len = sum(len) return(len) #返回此条旅行路径总距离}#根据每条染色体的分数计算权重,并以此抽样roller = function(scores,k){ scores = max(scores)-scores+1 props = scores/sum(scores) N = length(scores) mxind = which.max(scores)#保留最优染色体 ans = sample((1:N)[-mxind],k-1,replace = F,prob = props[-mxind]) return(c(mxind,ans))}#种群中的繁殖过程crossEvolve = function(i,nGroup,crossGroup,prop){ a = nGroup[i,] b = crossGroup[i,] n = length(a) m = max(1,trunc(n*prop)) tmpa = a st = sample(1:n,1) ind = st:(st+m) #indication 指示、索引 if (st+m>n) { bind = which(ind>n) ind[bind] = ind[bind] %% n + 1 } cross = intersect(b,a[ind]) tmpa[ind] = cross return(tmpa)}#染色体的自我变异selfVariation = function(dna,prop){ n = length(dna) pos = which(runif(n)<prop) if (length(pos)==0) return(dna) pos = sample(pos,1) newind = sample((1:n)[-pos],1) if (pos>newind) { tmp = dna[newind:n] tmp = tmp[-(pos-newind+1)] dna[newind] = dna[pos] dna[(newind+1):n] = tmp } else { tmp = dna[1:newind] tmp = tmp[-pos] dna[newind] = dna[pos] dna[1:(newind-1)] = tmp } return(dna)}#以某条染色体代表的解做图drawIt = function(dots,dna,xlab=NULL,ylab=NULL,main=NULL,sub=NULL,col=NULL){#win.graph(width=800, height=800, pointsize=12) x = dots[[1]] y = dots[[2]] z = dots[[4]] cols=dots[[5]]+10 if (is.null(main)) { scores = calcScores(dna,dots[[3]])#画地图 plot(border,border="#BBFFFF",col="#FF7F00",ylim = c(18, 54), panel.first = grid(), main=paste("总路程",scores,"公里"),xlab="经度",ylab="纬度",sub=paste('优化结束-第',sub,'代'));#增加省会城市坐标点 points(x,y,pch=19,col=cols) text(x,y,z,pos=1,cex=0.5) n = length(dna) for (i in 1:(n-1)){ Sys.sleep(0.3) lines(x[dna[i:(i+1)]],y[dna[i:(i+1)]],col="#00FFFF",lwd=3)} lines(x[dna[c(n,1)]],y[dna[c(n,1)]],col="#00FFFF",lwd=3) } else{#画地图plot(border,border="#BBFFFF",col="#FF7F00",ylim = c(18, 54),main=paste("总路程",main,"公里"), xlab="经度",ylab="纬度",sub=paste('第',sub,'代'))points(x,y,pch=19,col=cols)if(sub==1)Sys.sleep(5)else{ n = length(dna) for (i in 1:(n-1)) lines(x[dna[i:(i+1)]],y[dna[i:(i+1)]],col="#00FFFF",lwd=3) lines(x[dna[c(n,1)]],y[dna[c(n,1)]],col="#00FFFF",lwd=3)}}}#Genetic Algorithm for Traveller Salesman ProblemGA4TSP = function(dots,initDNA=NULL,N,cp,vp,maxIter,maxStay,maxElite,drawing){ disM = dots[[3]] n = nrow(disM) if (N %%2 >0) N = N+1 Group = t(sapply(rep(n,N),rndDNA)) if (!is.null(initDNA)) Group[1,]=initDNA maxL = UpperBound(disM) stopFlag = FALSE iterCount = 1 stayCount = 0 allBest = maxL eliteBest = maxL elite = mat.or.vec(maxElite,n) elitecount = 0 eracount = 0 LastEra = maxL outputRecorder = NULL GenerationRecorder = NULL showScore=FALSE eliteInto=FALSE #初始化结束 while (!stopFlag) { cat('Generation:',iterCount,'Era:',eracount,'Elite:',elitecount) scores = apply(Group,1,calcScores,disM) bestScore = min(scores) mind = which.min(scores) bestDNA = Group[mind,]#记录最佳染色体 #更新时代、精英的信息 if (bestScore<eliteBest) { stayCount = 0 eliteBest = bestScore eliteDNA = bestDNA if (eliteBest<allBest) { allBest = eliteBest allDNA = eliteDNA outputRecorder = rbind(outputRecorder,allDNA) GenerationRecorder = c(GenerationRecorder,iterCount) if (drawing) drawIt(dots,allDNA,main=as.character(allBest),sub=iterCount) } } else stayCount = stayCount+1 if (stayCount == maxStay) { stayCount = 0 eliteBest = maxL elitecount = elitecount+1 elite[elitecount,] = eliteDNA scores = apply(Group,1,calcScores,disM) a = which(scores == min(scores)) nind = sample((1:N)[-a],length(a)) Group[a,] = Group[nind,] eliteInto =TRUE scores = apply(Group,1,calcScores,disM) bestScore = min(scores) mind = which.min(scores) bestDNA = Group[mind,] } if (elitecount==maxElite) { Group[1:elitecount,]=elite elite = mat.or.vec(maxElite,n) elitecount = 0 stayCount = 0 eliteBest = maxL eracount = eracount+1 maxStay = maxStay + 20 showScore=TRUE scores = apply(Group,1,calcScores,disM) bestScore = min(scores) mind = which.min(scores) bestDNA = Group[mind,] } #对种群计算分数,产生繁殖与变异 succind = roller(scores,N/2) nGroup = Group[succind,] #取最优和一半高权重优秀基因 crossind = sample(succind,N/2) crossGroup = Group[crossind,] #取最优和一半高权重优秀基因,打乱顺序 crossans = t(sapply(1:(N/2),crossEvolve,nGroup,crossGroup,cp)) crossGroup = rbind(nGroup,crossans) Group = t(apply(crossGroup,1,selfVariation,vp)) if (eliteInto) eliteInto = FALSE else Group[1,] = bestDNA stopFlag = (iterCount>=maxIter) iterCount = iterCount+1 cat(' Best:',bestScore,'All:',allBest,'Stay:',paste(stayCount,'/',maxStay,sep=''),'\n') if (showScore) { scores = apply(Group,1,calcScores,disM) show(scores[1:(N/2)]) show(scores[(N/2+1):(N)]) #Sys.sleep(1) showScore=FALSE } } if (drawing) drawIt(dots,allDNA,sub=maxIter) return(list(DNA = outputRecorder,Generation=GenerationRecorder))}#生成初始染色体CreatDNA = function(data,i){hc=hclust(dist(data[,1:2]))data\(col</code><code class="python keyword">=</code><code class="python plain">cutree(hc, k </code><code class="python keyword">=</code> <code class="python plain">i) </code><code class="python comments">#k = 1 is trivial</code></div><div class="line number250 index249 alt1"><code class="python plain">INITDNA</code><code class="python keyword">=</code><code class="python plain">data[order(data\)col),]rownames(INITDNA)=NULL return(INITDNA) }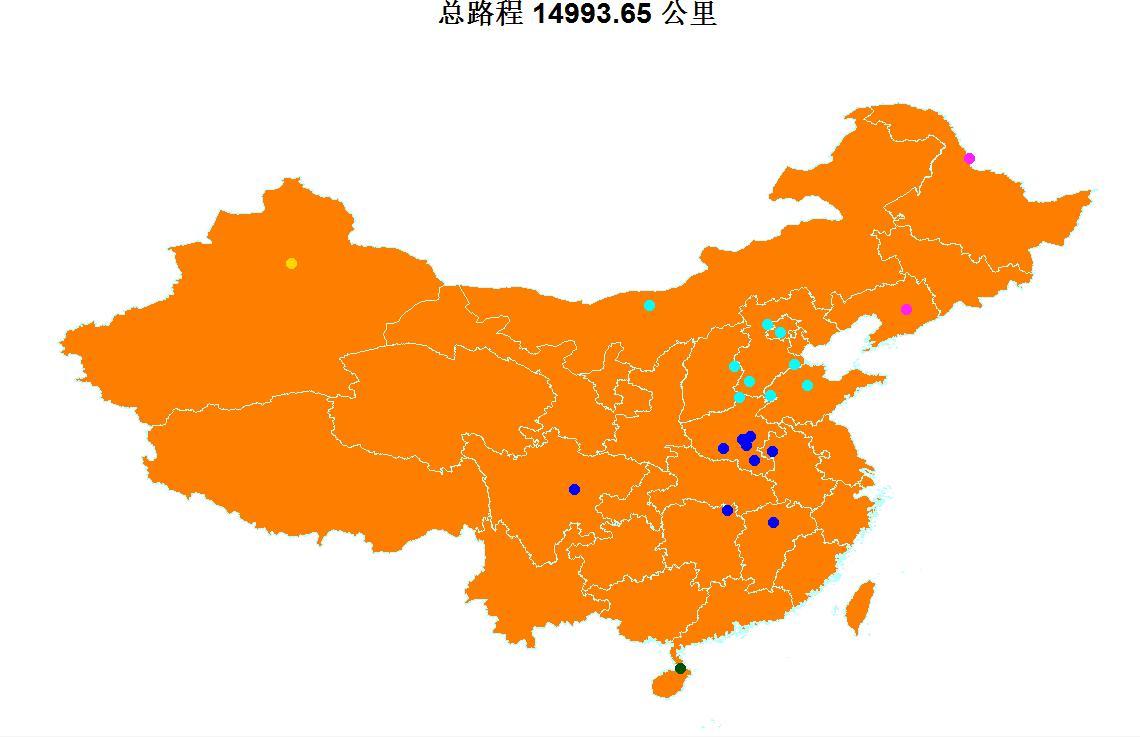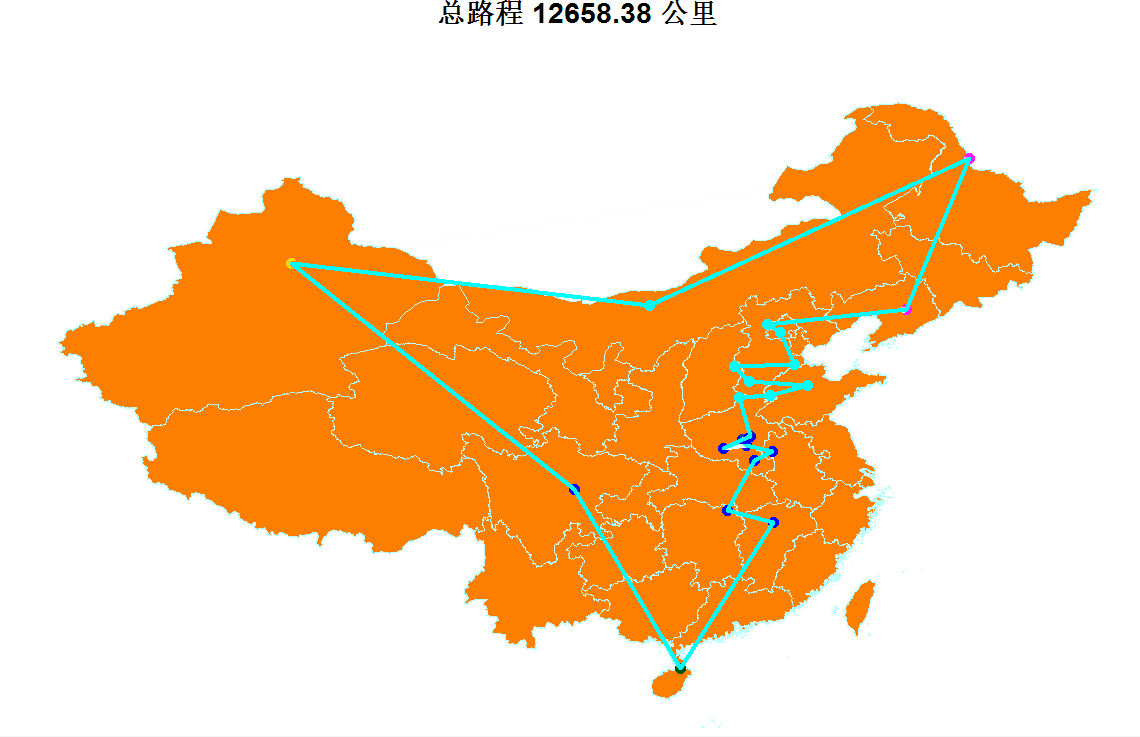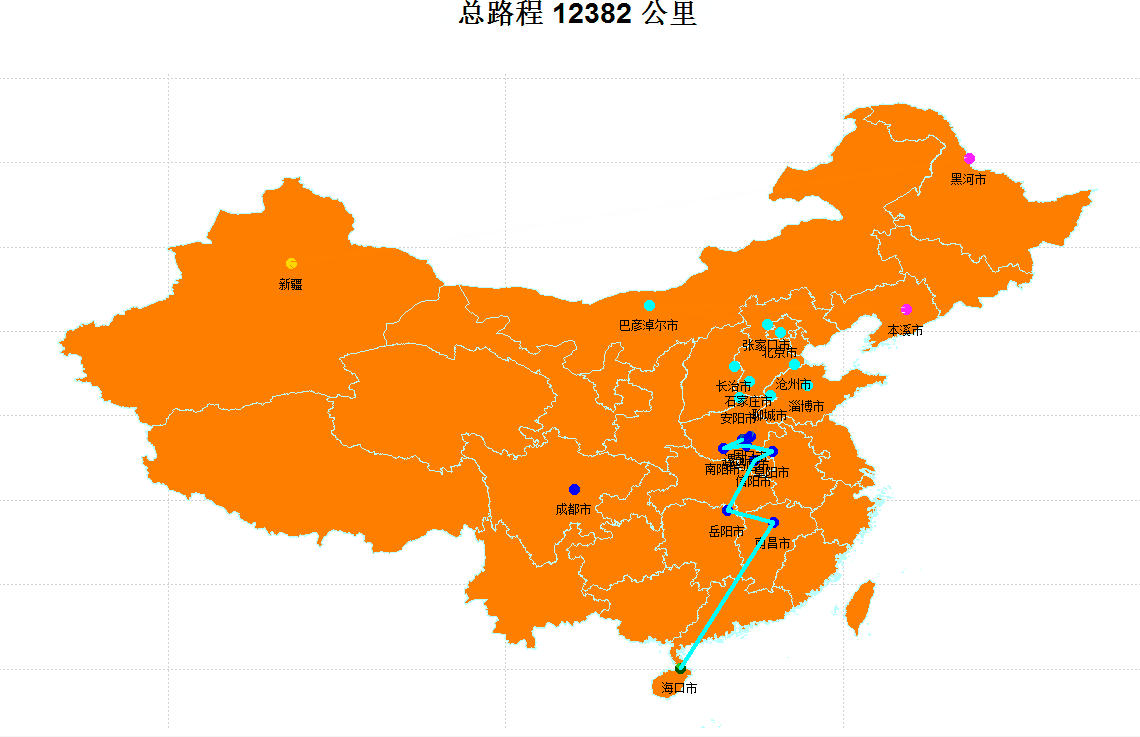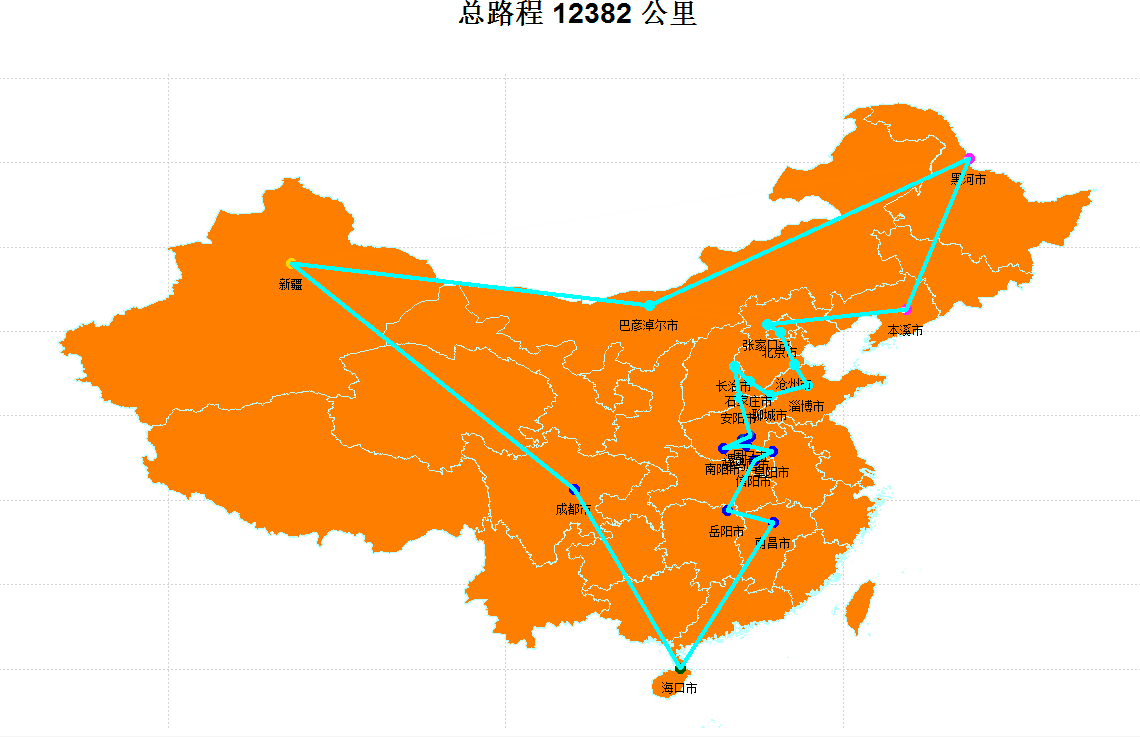



 浙公网安备 33010602011771号
浙公网安备 33010602011771号