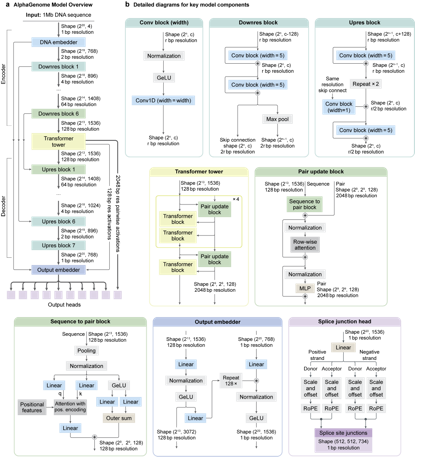
【AI生物技术前沿】AlphaGenome
原文:x.com/s6juncheng/status/1937873856212996257
作者:Jun Cheng
Excited to share #AlphaGenome, a start of our AlphaGenome named journey to decipher the regulatory genome! The model matches or exceeds top-performing external models on 24 out of 26 variant evaluations, across a wide range of biological modalities.

Mistakes in RNA splicing are a frequent cause of diseases. For the first time, we've built a single model that unifies the prediction of RNA-seq coverage, splice sites, their usage, AND the specific junctions they form, providing a more complete picture of splicing outcomes.
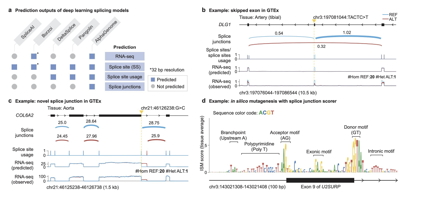
This more complete view can help us understand the complex consequences of genetic variants on RNA splicing. It is SOTA on 6 out of 7 splicing variant benchmarks, including predicting splicing variant pathogenicity from ClinVar, sQTL causality, and GTEx splicing outliers.

One of the most exciting parts of our #AlphaGenome work is the ability to directly predict splice junctions from sequence and also use it for variant effect prediction. This is enabled by modeling the competition between splice sites and junction supporting reads.
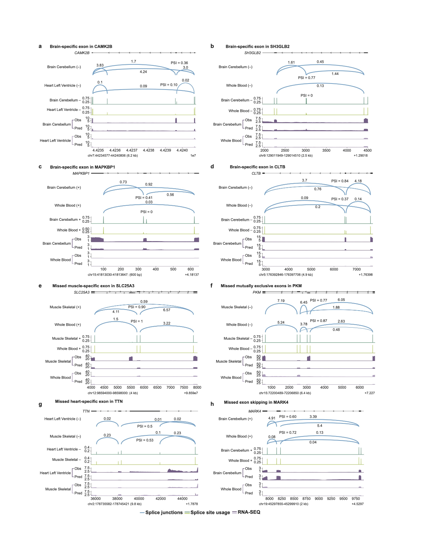
The model predicts splicing for 367 human tissues or cell types on both strands. Tissue-specific splicing was predicted with some success, but in general this is still a hard problem. Here we show some examples with mixed results.
Having worked on RNA splicing models for years, this is a significant milestone for me. It is a real privilege to work with an extremely talented team. With the new API, I’m excited to see what researchers can find with #AlphaGenome.
The model source code and weights will also be provided upon final publication.


 浙公网安备 33010602011771号
浙公网安备 33010602011771号