题目
DNA sequence
*Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Total Submission(s): 7590 Accepted Submission(s): 3354
*Problem Description
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence of it.
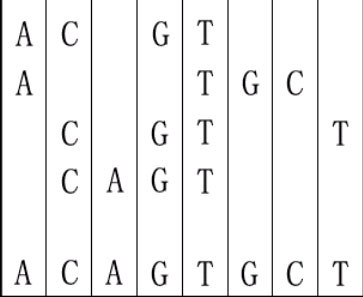
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
Input
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
Output
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
Sample Input
1 4 ACGT ATGC CGTT CAGTSample Output
8Author
LL
Source
思路 (IDA* 迭代式深搜)
迭代式深搜的目的是为了减少空间上的浪费,众所周知BFS队列会提升空间复杂度, IDA*是限制深度
的深搜,对每个深度进行枚举,若达到上限深度则停止。
对于本题,最大深度是所有DNA序列之和,最小深度是从所有字符中最大的开始,然后进行dfs,若当前匹配需要的最小深度大于当前的限制深度,那么我们直接return,当前匹配的最小深度可以通过计算每个串的匹配位置来得到,这样我们就可以类似于BFS的进行每一层的搜索
Code
#include <iostream>
#include <vector>
#include <algorithm>
#include <cstring>
using namespace std;
const int N = 10;
string ex = "ATCG";
// pos数组负责记录当前串的匹配位置
int n, deep, pos[N], ans;
vector<string> a;
string now;
bool flag;
int cal(int pos[]) {
int mx = 0;
for (int i = 0; i < a.size(); i ++) {
int len = (int) a[i].length();
// 最小所需深度=max(当前串的匹配位置减去其长度,即还所需匹配的字符)
mx = max(mx, len - pos[i]);
}
return mx;
}
void dfs(int u, int pos[]) {
if (u > deep) return;
// cout << u << endl;
if (flag) return;
// 计算所需最小深度
int mx = cal(pos);
// 若当前需要的最小深度大于当前深度或需要的深度为0,则找到答案
if (u + mx > deep || !mx) {
if (!mx) ans = u;
return;
}
// t为更新后的pos
int t[N];
// 枚举ACTG进行DFS
for (int i = 0; i <= 3; i ++) {
bool check = false;
for (int j = 0; j < a.size(); j ++) {
t[j] = pos[j];
// 若当前的pos位小于当前匹配串的大小,并且和当前串所对应字符相同
if (pos[j] < a[j].size() && ex[i] == a[j][pos[j]]) {
t[j] = pos[j] + 1;
check = true;
}
}
if (check) dfs(u + 1, t);
if (flag) return;
}
}
int main() {
string t;
int _;
cin >> _;
while (_ --) {
deep = 0;
a.clear();
cin >> n;
for (int i = 0; i < n; i ++) {
cin >> t;
a.push_back(t);
// 记录最小所需深度
deep = max(deep, (int) t.size());
}
ans = -1;
// ida*枚举每个深度进行DFS,相当于DFS版本的宽搜
while(true) {
memset(pos, 0, sizeof pos);
dfs(0, pos);
if (ans != -1) {
cout << ans << endl;
break;
}
deep ++;
}
}
}


 浙公网安备 33010602011771号
浙公网安备 33010602011771号