This is a short introduction to pandas, geared mainly for new users. You can see more complex recipes in the Cookbook
Customarily, we import as follows:
In [1]: import pandas as pd
In [2]: import numpy as np
In [3]: import matplotlib.pyplot as plt
Object Creation
See the Data Structure Intro section
------------------------------------------------------------------------------------------------------------
Intro to Data Structures
We’ll start with a quick, non-comprehensive overview of the fundamental data structures in pandas to get you started. The fundamental behavior about data types, indexing, and axis labeling / alignment apply across all of the objects. To get started, import numpy and load pandas into your namespace:
In [1]: import numpy as np
In [2]: import pandas as pd
Here is a basic tenet to keep in mind: data alignment is intrinsic. The link between labels and data will not be broken unless done so explicitly by you.
We’ll give a brief intro to the data structures, then consider all of the broad categories of functionality and methods in separate sections.
Series
Series is a one-dimensional labeled array capable of holding any data type (integers, strings, floating point numbers, Python objects, etc.). The axis labels are collectively referred to as the index. The basic method to create a Series is to call:
>>> s = pd.Series(data, index=index)
Here, data can be many different things:
- a Python dict
- an ndarray
- a scalar value (like 5)
The passed index is a list of axis labels. Thus, this separates into a few cases depending on what data is:
From ndarray
If data is an ndarray, index must be the same length as data. If no index is passed, one will be created having values [0, ..., len(data) - 1].
In [3]: s = pd.Series(np.random.randn(5), index=['a', 'b', 'c', 'd', 'e'])
In [4]: s
Out[4]:
a 0.4691
b -0.2829
c -1.5091
d -1.1356
e 1.2121
dtype: float64
In [5]: s.index
Out[5]: Index(['a', 'b', 'c', 'd', 'e'], dtype='object')
In [6]: pd.Series(np.random.randn(5))
Out[6]:
0 -0.1732
1 0.1192
2 -1.0442
3 -0.8618
4 -2.1046
dtype: float64
Note
pandas supports non-unique index values. If an operation that does not support duplicate index values is attempted, an exception will be raised at that time. The reason for being lazy is nearly all performance-based (there are many instances in computations, like parts of GroupBy, where the index is not used).
From dict
If data is a dict, if index is passed the values in data corresponding to the labels in the index will be pulled out. Otherwise, an index will be constructed from the sorted keys of the dict, if possible.
In [7]: d = {'a' : 0., 'b' : 1., 'c' : 2.}
In [8]: pd.Series(d)
Out[8]:
a 0.0
b 1.0
c 2.0
dtype: float64
In [9]: pd.Series(d, index=['b', 'c', 'd', 'a'])
Out[9]:
b 1.0
c 2.0
d NaN
a 0.0
dtype: float64
Note
NaN (not a number) is the standard missing data marker used in pandas
From scalar value If data is a scalar value, an index must be provided. The value will be repeated to match the length of index
In [10]: pd.Series(5., index=['a', 'b', 'c', 'd', 'e'])
Out[10]:
a 5.0
b 5.0
c 5.0
d 5.0
e 5.0
dtype: float64
Series is ndarray-like
Series acts very similarly to a ndarray, and is a valid argument to most NumPy functions. However, things like slicing also slice the index.
In [11]: s[0]
Out[11]: 0.46911229990718628
In [12]: s[:3]
Out[12]:
a 0.4691
b -0.2829
c -1.5091
dtype: float64
In [13]: s[s > s.median()] #先排序,后中值
Out[13]:
a 0.4691
e 1.2121
dtype: float64
In [14]: s[[4, 3, 1]]
Out[14]:
e 1.2121
d -1.1356
b -0.2829
dtype: float64
In [15]: np.exp(s)
Out[15]:
a 1.5986
b 0.7536
c 0.2211
d 0.3212
e 3.3606
dtype: float64
We will address array-based indexing in a separate section.
Series is dict-like
A Series is like a fixed-size dict in that you can get and set values by index label:
In [16]: s['a']
Out[16]: 0.46911229990718628
In [17]: s['e'] = 12.
In [18]: s
Out[18]:
a 0.4691
b -0.2829
c -1.5091
d -1.1356
e 12.0000
dtype: float64
In [19]: 'e' in s
Out[19]: True
In [20]: 'f' in s
Out[20]: False
If a label is not contained, an exception is raised:
>>> s['f']
KeyError: 'f'
Using the get method, a missing label will return None or specified default:
In [21]: s.get('f')
In [22]: s.get('f', np.nan)
Out[22]: nan
See also the section on attribute access.
Vectorized operations and label alignment with Series
When doing data analysis, as with raw NumPy arrays looping through Series value-by-value is usually not necessary. Series can also be passed into most NumPy methods expecting an ndarray.
In [23]: s + s
Out[23]:
a 0.9382
b -0.5657
c -3.0181
d -2.2713
e 24.0000
dtype: float64
In [24]: s * 2
Out[24]:
a 0.9382
b -0.5657
c -3.0181
d -2.2713
e 24.0000
dtype: float64
In [25]: np.exp(s)
Out[25]:
a 1.5986
b 0.7536
c 0.2211
d 0.3212
e 162754.7914
dtype: float64
A key difference between Series and ndarray is that operations between Series automatically align the data based on label. Thus, you can write computations without giving consideration to whether the Series involved have the same labels.
In [26]: s[1:] + s[:-1]
Out[26]:
a NaN
b -0.5657
c -3.0181
d -2.2713
e NaN
dtype: float64
The result of an operation between unaligned Series will have the union of the indexes involved. If a label is not found in one Series or the other, the result will be marked as missing NaN. Being able to write code without doing any explicit data alignment grants immense freedom and flexibility in interactive data analysis and research. The integrated data alignment features of the pandas data structures set pandas apart from the majority of related tools for working with labeled data.
Note
In general, we chose to make the default result of operations between differently indexed objects yield the union of the indexes in order to avoid loss of information. Having an index label, though the data is missing, is typically important information as part of a computation. You of course have the option of dropping labels with missing data via thedropna function.
Name attribute
Series can also have a name attribute:
In [27]: s = pd.Series(np.random.randn(5), name='something')
In [28]: s
Out[28]:
0 -0.4949
1 1.0718
2 0.7216
3 -0.7068
4 -1.0396
Name: something, dtype: float64
In [29]: s.name
Out[29]: 'something'
The Series name will be assigned automatically in many cases, in particular when taking 1D slices of DataFrame as you will see below.
New in version 0.18.0.
You can rename a Series with the pandas.Series.rename() method.
In [30]: s2 = s.rename("different")
In [31]: s2.name
Out[31]: 'different'
Note that s and s2 refer to different objects.
DataFrame
DataFrame is a 2-dimensional labeled data structure with columns of potentially different types. You can think of it like a spreadsheet or SQL table, or a dict of Series objects. It is generally the most commonly used pandas object. Like Series, DataFrame accepts many different kinds of input:
- Dict of 1D ndarrays, lists, dicts, or Series
- 2-D numpy.ndarray
- Structured or record ndarray
- A
Series- Another
DataFrame
Along with the data, you can optionally pass index (row labels) and columns (column labels) arguments. If you pass an index and / or columns, you are guaranteeing the index and / or columns of the resulting DataFrame. Thus, a dict of Series plus a specific index will discard all data not matching up to the passed index.
If axis labels are not passed, they will be constructed from the input data based on common sense rules.
From dict of Series or dicts
The result index will be the union of the indexes of the various Series. If there are any nested dicts, these will be first converted to Series. If no columns are passed, the columns will be the sorted list of dict keys.
In [32]: d = {'one' : pd.Series([1., 2., 3.], index=['a', 'b', 'c']),
....: 'two' : pd.Series([1., 2., 3., 4.], index=['a', 'b', 'c', 'd'])}
....:
In [33]: df = pd.DataFrame(d)
In [34]: df
Out[34]:
one two
a 1.0 1.0
b 2.0 2.0
c 3.0 3.0
d NaN 4.0
In [35]: pd.DataFrame(d, index=['d', 'b', 'a'])
Out[35]:
one two
d NaN 4.0
b 2.0 2.0
a 1.0 1.0
In [36]: pd.DataFrame(d, index=['d', 'b', 'a'], columns=['two', 'three'])
Out[36]:
two three
d 4.0 NaN
b 2.0 NaN
a 1.0 NaN
The row and column labels can be accessed respectively by accessing the index and columns attributes:
Note
When a particular set of columns is passed along with a dict of data, the passed columns override the keys in the dict.
In [37]: df.index
Out[37]: Index(['a', 'b', 'c', 'd'], dtype='object')
In [38]: df.columns
Out[38]: Index(['one', 'two'], dtype='object')
From dict of ndarrays / lists
The ndarrays must all be the same length. If an index is passed, it must clearly also be the same length as the arrays. If no index is passed, the result will be range(n), where n is the array length.
In [39]: d = {'one' : [1., 2., 3., 4.],
....: 'two' : [4., 3., 2., 1.]}
....:
In [40]: pd.DataFrame(d)
Out[40]:
one two
0 1.0 4.0
1 2.0 3.0
2 3.0 2.0
3 4.0 1.0
In [41]: pd.DataFrame(d, index=['a', 'b', 'c', 'd'])
Out[41]:
one two
a 1.0 4.0
b 2.0 3.0
c 3.0 2.0
d 4.0 1.0
From structured or record array
This case is handled identically to a dict of arrays.
In [42]: data = np.zeros((2,), dtype=[('A', 'i4'),('B', 'f4'),('C', 'a10')]) #建立数据结构
In [43]: data[:] = [(1,2.,'Hello'), (2,3.,"World")] #填数据
In [44]: pd.DataFrame(data)
Out[44]:
A B C
0 1 2.0 b'Hello'
1 2 3.0 b'World'
In [45]: pd.DataFrame(data, index=['first', 'second'])
Out[45]:
A B C
first 1 2.0 b'Hello'
second 2 3.0 b'World'
In [46]: pd.DataFrame(data, columns=['C', 'A', 'B'])
Out[46]:
C A B
0 b'Hello' 1 2.0
1 b'World' 2 3.0
Note
DataFrame is not intended to work exactly like a 2-dimensional NumPy ndarray.
From a list of dicts
In [47]: data2 = [{'a': 1, 'b': 2}, {'a': 5, 'b': 10, 'c': 20}]
In [48]: pd.DataFrame(data2)
Out[48]:
a b c
0 1 2 NaN
1 5 10 20.0
In [49]: pd.DataFrame(data2, index=['first', 'second'])
Out[49]:
a b c
first 1 2 NaN
second 5 10 20.0
In [50]: pd.DataFrame(data2, columns=['a', 'b'])
Out[50]:
a b
0 1 2
1 5 10
From a dict of tuples
You can automatically create a multi-indexed frame by passing a tuples dictionary
In [51]: pd.DataFrame({('a', 'b'): {('A', 'B'): 1, ('A', 'C'): 2},
....: ('a', 'a'): {('A', 'C'): 3, ('A', 'B'): 4},
....: ('a', 'c'): {('A', 'B'): 5, ('A', 'C'): 6},
....: ('b', 'a'): {('A', 'C'): 7, ('A', 'B'): 8},
....: ('b', 'b'): {('A', 'D'): 9, ('A', 'B'): 10}})
....:
Out[51]:
a b
a b c a b
A B 4.0 1.0 5.0 8.0 10.0
C 3.0 2.0 6.0 7.0 NaN
D NaN NaN NaN NaN 9.0
![]()
-----待更
From a Series
The result will be a DataFrame with the same index as the input Series, and with one column whose name is the original name of the Series (only if no other column name provided).
Missing Data
Much more will be said on this topic in the Missing data section. To construct a DataFrame with missing data, use np.nan for those values which are missing. Alternatively, you may pass a numpy.MaskedArray as the data argument to the DataFrame constructor, and its masked entries will be considered missing.
Alternate Constructors
DataFrame.from_dict
DataFrame.from_dict takes a dict of dicts or a dict of array-like sequences and returns a DataFrame. It operates like the DataFrame constructor except for the orient parameter which is 'columns' by default, but which can be set to 'index' in order to use the dict keys as row labels.
DataFrame.from_records
DataFrame.from_records takes a list of tuples or an ndarray with structured dtype. Works analogously to the normal DataFrame constructor, except that index maybe be a specific field of the structured dtype to use as the index. For example:
In [52]: data
Out[52]:
array([(1, 2., b'Hello'), (2, 3., b'World')],
dtype=[('A', '<i4'), ('B', '<f4'), ('C', 'S10')])
In [53]: pd.DataFrame.from_records(data, index='C')
Out[53]:
A B
C
b'Hello' 1 2.0
b'World' 2 3.0
DataFrame.from_items
DataFrame.from_items works analogously to the form of the dict constructor that takes a sequence of (key,value) pairs, where the keys are column (or row, in the case of orient='index') names, and the value are the column values (or row values). This can be useful for constructing a DataFrame with the columns in a particular order without having to pass an explicit list of columns:
In [54]: pd.DataFrame.from_items([('A', [1, 2, 3]), ('B', [4, 5, 6])])
Out[54]:
A B
0 1 4
1 2 5
2 3 6
If you pass orient='index', the keys will be the row labels. But in this case you must also pass the desired column names:
In [55]: pd.DataFrame.from_items([('A', [1, 2, 3]), ('B', [4, 5, 6])],
....: orient='index', columns=['one', 'two', 'three'])
....:
Out[55]:
one two three
A 1 2 3
B 4 5 6
Column selection, addition, deletion
You can treat a DataFrame semantically like a dict of like-indexed Series objects. Getting, setting, and deleting columns works with the same syntax as the analogous dict operations:
In [56]: df['one']
Out[56]:
a 1.0
b 2.0
c 3.0
d NaN
Name: one, dtype: float64
In [57]: df['three'] = df['one'] * df['two']
In [58]: df['flag'] = df['one'] > 2
In [59]: df
Out[59]:
one two three flag
a 1.0 1.0 1.0 False
b 2.0 2.0 4.0 False
c 3.0 3.0 9.0 True
d NaN 4.0 NaN False
Columns can be deleted or popped like with a dict:
In [60]: del df['two']
In [61]: three = df.pop('three')
In [62]: df
Out[62]:
one flag
a 1.0 False
b 2.0 False
c 3.0 True
d NaN False
When inserting a scalar value, it will naturally be propagated to fill the column:
In [63]: df['foo'] = 'bar'
In [64]: df
Out[64]:
one flag foo
a 1.0 False bar
b 2.0 False bar
c 3.0 True bar
d NaN False bar
When inserting a Series that does not have the same index as the DataFrame, it will be conformed to the DataFrame’s index:
In [65]: df['one_trunc'] = df['one'][:2]
In [66]: df
Out[66]:
one flag foo one_trunc
a 1.0 False bar 1.0
b 2.0 False bar 2.0
c 3.0 True bar NaN
d NaN False bar NaN
You can insert raw ndarrays but their length must match the length of the DataFrame’s index.
By default, columns get inserted at the end. The insert function is available to insert at a particular location in the columns:
In [67]: df.insert(1, 'bar', df['one'])
In [68]: df
Out[68]:
one bar flag foo one_trunc
a 1.0 1.0 False bar 1.0
b 2.0 2.0 False bar 2.0
c 3.0 3.0 True bar NaN
d NaN NaN False bar NaN
Assigning New Columns in Method Chains
Inspired by dplyr’s mutate verb, DataFrame has an assign() method that allows you to easily create new columns that are potentially derived from existing columns.
In [69]: iris = pd.read_csv('data/iris.data')
In [70]: iris.head()
Out[70]:
SepalLength SepalWidth PetalLength PetalWidth Name
0 5.1 3.5 1.4 0.2 Iris-setosa
1 4.9 3.0 1.4 0.2 Iris-setosa
2 4.7 3.2 1.3 0.2 Iris-setosa
3 4.6 3.1 1.5 0.2 Iris-setosa
4 5.0 3.6 1.4 0.2 Iris-setosa
In [71]: (iris.assign(sepal_ratio = iris['SepalWidth'] / iris['SepalLength'])
....: .head())
....:
Out[71]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.6863
1 4.9 3.0 1.4 0.2 Iris-setosa 0.6122
2 4.7 3.2 1.3 0.2 Iris-setosa 0.6809
3 4.6 3.1 1.5 0.2 Iris-setosa 0.6739
4 5.0 3.6 1.4 0.2 Iris-setosa 0.7200
Above was an example of inserting a precomputed value. We can also pass in a function of one argument to be evalutated on the DataFrame being assigned to.
In [72]: iris.assign(sepal_ratio = lambda x: (x['SepalWidth'] /
....: x['SepalLength'])).head()
....:
Out[72]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.6863
1 4.9 3.0 1.4 0.2 Iris-setosa 0.6122
2 4.7 3.2 1.3 0.2 Iris-setosa 0.6809
3 4.6 3.1 1.5 0.2 Iris-setosa 0.6739
4 5.0 3.6 1.4 0.2 Iris-setosa 0.7200
assign always returns a copy of the data, leaving the original DataFrame untouched.
Passing a callable, as opposed to an actual value to be inserted, is useful when you don’t have a reference to the DataFrame at hand. This is common when using assign in chains of operations. For example, we can limit the DataFrame to just those observations with a Sepal Length greater than 5, calculate the ratio, and plot:
In [73]: (iris.query('SepalLength > 5')
....: .assign(SepalRatio = lambda x: x.SepalWidth / x.SepalLength,
....: PetalRatio = lambda x: x.PetalWidth / x.PetalLength)
....: .plot(kind='scatter', x='SepalRatio', y='PetalRatio'))
....:
Out[73]: <matplotlib.axes._subplots.AxesSubplot at 0x122d7fba8>
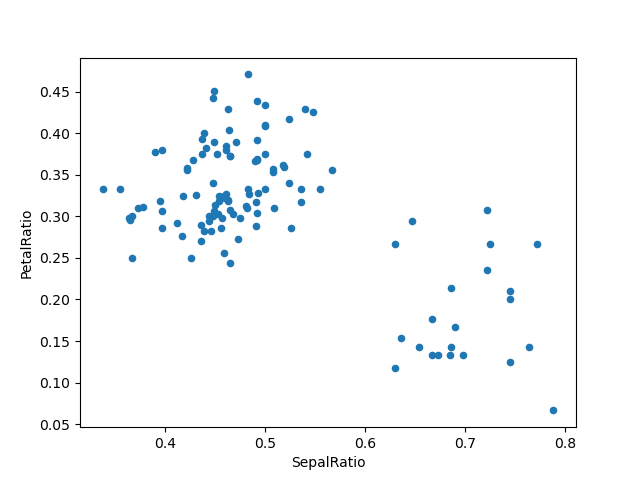
Since a function is passed in, the function is computed on the DataFrame being assigned to. Importantly, this is the DataFrame that’s been filtered to those rows with sepal length greater than 5. The filtering happens first, and then the ratio calculations. This is an example where we didn’t have a reference to the filtered DataFrame available.
The function signature for assign is simply **kwargs. The keys are the column names for the new fields, and the values are either a value to be inserted (for example, a Series or NumPy array), or a function of one argument to be called on the DataFrame. A copy of the original DataFrame is returned, with the new values inserted.
Warning
Since the function signature of assign is **kwargs, a dictionary, the order of the new columns in the resulting DataFrame cannot be guaranteed to match the order you pass in. To make things predictable, items are inserted alphabetically (by key) at the end of the DataFrame.
All expressions are computed first, and then assigned. So you can’t refer to another column being assigned in the same call to assign. For example:
In [74]: # Don't do this, bad reference to `C` df.assign(C = lambda x: x['A'] + x['B'], D = lambda x: x['A'] + x['C']) In [2]: # Instead, break it into two assigns (df.assign(C = lambda x: x['A'] + x['B']) .assign(D = lambda x: x['A'] + x['C']))
Indexing / Selection
The basics of indexing are as follows:
| Operation | Syntax | Result |
|---|---|---|
| Select column | df[col] |
Series |
| Select row by label | df.loc[label] |
Series |
| Select row by integer location | df.iloc[loc] |
Series |
| Slice rows | df[5:10] |
DataFrame |
| Select rows by boolean vector | df[bool_vec] |
DataFrame |
Row selection, for example, returns a Series whose index is the columns of the DataFrame:
In [75]: df.loc['b']
Out[75]:
one 2
bar 2
flag False
foo bar
one_trunc 2
Name: b, dtype: object
In [76]: df.iloc[2]
Out[76]:
one 3
bar 3
flag True
foo bar
one_trunc NaN
Name: c, dtype: object
For a more exhaustive treatment of more sophisticated label-based indexing and slicing, see the section on indexing. We will address the fundamentals of reindexing / conforming to new sets of labels in the section on reindexing.
Data alignment and arithmetic
Data alignment between DataFrame objects automatically align on both the columns and the index (row labels). Again, the resulting object will have the union of the column and row labels.
In [77]: df = pd.DataFrame(np.random.randn(10, 4), columns=['A', 'B', 'C', 'D'])
In [78]: df2 = pd.DataFrame(np.random.randn(7, 3), columns=['A', 'B', 'C'])
In [79]: df + df2
Out[79]:
A B C D
0 0.0457 -0.0141 1.3809 NaN
1 -0.9554 -1.5010 0.0372 NaN
2 -0.6627 1.5348 -0.8597 NaN
3 -2.4529 1.2373 -0.1337 NaN
4 1.4145 1.9517 -2.3204 NaN
5 -0.4949 -1.6497 -1.0846 NaN
6 -1.0476 -0.7486 -0.8055 NaN
7 NaN NaN NaN NaN
8 NaN NaN NaN NaN
9 NaN NaN NaN NaN
When doing an operation between DataFrame and Series, the default behavior is to align the Series index on the DataFrame columns, thus broadcasting row-wise. For example:
In [80]: df - df.iloc[0]
Out[80]:
A B C D
0 0.0000 0.0000 0.0000 0.0000
1 -1.3593 -0.2487 -0.4534 -1.7547
2 0.2531 0.8297 0.0100 -1.9912
3 -1.3111 0.0543 -1.7249 -1.6205
4 0.5730 1.5007 -0.6761 1.3673
5 -1.7412 0.7820 -1.2416 -2.0531
6 -1.2408 -0.8696 -0.1533 0.0004
7 -0.7439 0.4110 -0.9296 -0.2824
8 -1.1949 1.3207 0.2382 -1.4826
9 2.2938 1.8562 0.7733 -1.4465
In the special case of working with time series data, and the DataFrame index also contains dates, the broadcasting will be column-wise:
In [81]: index = pd.date_range('1/1/2000', periods=8) ????????????????????????
In [82]: df = pd.DataFrame(np.random.randn(8, 3), index=index, columns=list('ABC'))
In [83]: df
Out[83]:
A B C
2000-01-01 -1.2268 0.7698 -1.2812
2000-01-02 -0.7277 -0.1213 -0.0979
2000-01-03 0.6958 0.3417 0.9597
2000-01-04 -1.1103 -0.6200 0.1497
2000-01-05 -0.7323 0.6877 0.1764
2000-01-06 0.4033 -0.1550 0.3016
2000-01-07 -2.1799 -1.3698 -0.9542
2000-01-08 1.4627 -1.7432 -0.8266
In [84]: type(df['A'])
Out[84]: pandas.core.series.Series
In [85]: df - df['A']
Out[85]:
2000-01-01 00:00:00 2000-01-02 00:00:00 2000-01-03 00:00:00 \
2000-01-01 NaN NaN NaN
2000-01-02 NaN NaN NaN
2000-01-03 NaN NaN NaN
2000-01-04 NaN NaN NaN
2000-01-05 NaN NaN NaN
2000-01-06 NaN NaN NaN
2000-01-07 NaN NaN NaN
2000-01-08 NaN NaN NaN
2000-01-04 00:00:00 ... 2000-01-08 00:00:00 A B C
2000-01-01 NaN ... NaN NaN NaN NaN
2000-01-02 NaN ... NaN NaN NaN NaN
2000-01-03 NaN ... NaN NaN NaN NaN
2000-01-04 NaN ... NaN NaN NaN NaN
2000-01-05 NaN ... NaN NaN NaN NaN
2000-01-06 NaN ... NaN NaN NaN NaN
2000-01-07 NaN ... NaN NaN NaN NaN
2000-01-08 NaN ... NaN NaN NaN NaN
[8 rows x 11 columns]
Warning
df - df['A']
is now deprecated and will be removed in a future release. The preferred way to replicate this behavior is
df.sub(df['A'], axis=0)
For explicit control over the matching and broadcasting behavior, see the section on flexible binary operations.
Operations with scalars are just as you would expect:
In [86]: df * 5 + 2
Out[86]:
A B C
2000-01-01 -4.1341 5.8490 -4.4062
2000-01-02 -1.6385 1.3935 1.5106
2000-01-03 5.4789 3.7087 6.7986
2000-01-04 -3.5517 -1.0999 2.7487
2000-01-05 -1.6617 5.4387 2.8822
2000-01-06 4.0165 1.2252 3.5081
2000-01-07 -8.8993 -4.8492 -2.7710
2000-01-08 9.3135 -6.7158 -2.1330
In [87]: 1 / df
Out[87]:
A B C
2000-01-01 -0.8151 1.2990 -0.7805
2000-01-02 -1.3742 -8.2436 -10.2163
2000-01-03 1.4372 2.9262 1.0420
2000-01-04 -0.9006 -1.6130 6.6779
2000-01-05 -1.3655 1.4540 5.6675
2000-01-06 2.4795 -6.4537 3.3154
2000-01-07 -0.4587 -0.7300 -1.0480
2000-01-08 0.6837 -0.5737 -1.2098
In [88]: df ** 4
Out[88]:
A B C
2000-01-01 2.2653 0.3512 2.6948e+00
2000-01-02 0.2804 0.0002 9.1796e-05
2000-01-03 0.2344 0.0136 8.4838e-01
2000-01-04 1.5199 0.1477 5.0286e-04
2000-01-05 0.2876 0.2237 9.6924e-04
2000-01-06 0.0265 0.0006 8.2769e-03
2000-01-07 22.5795 3.5212 8.2903e-01
2000-01-08 4.5774 9.2332 4.6683e-01
Boolean operators work as well:
In [89]: df1 = pd.DataFrame({'a' : [1, 0, 1], 'b' : [0, 1, 1] }, dtype=bool)
In [90]: df2 = pd.DataFrame({'a' : [0, 1, 1], 'b' : [1, 1, 0] }, dtype=bool)
In [91]: df1 & df2 #与
Out[91]:
a b
0 False False
1 False True
2 True False
In [92]: df1 | df2 #或
Out[92]:
a b
0 True True
1 True True
2 True True
In [93]: df1 ^ df2 #异或
Out[93]:
a b
0 True True
1 True False
2 False True
In [94]: -df1 #非
Out[94]:
a b
0 False True
1 True False
2 False False
Transposing
To transpose, access the T attribute (also the transpose function), similar to an ndarray:
# only show the first 5 rows
In [95]: df[:5].T
Out[95]:
2000-01-01 2000-01-02 2000-01-03 2000-01-04 2000-01-05
A -1.2268 -0.7277 0.6958 -1.1103 -0.7323
B 0.7698 -0.1213 0.3417 -0.6200 0.6877
C -1.2812 -0.0979 0.9597 0.1497 0.1764
置换
转置,访问 T 属性(也 转置 功能), 类似于一个ndarray:
# only show the first 5 rows
In [95]: df[:5].T
Out[95]:
2000-01-01 2000-01-02 2000-01-03 2000-01-04 2000-01-05
A -1.2268 -0.7277 0.6958 -1.1103 -0.7323
B 0.7698 -0.1213 0.3417 -0.6200 0.6877
C -1.2812 -0.0979 0.9597 0.1497 0.1764
DataFrame NumPy函数的互用性
Elementwise NumPy所有日志、exp√,…)和各种其他NumPy功能 可以用于DataFrame上没有问题,假设数据是数字:
In [96]: np.exp(df)
Out[96]:
A B C
2000-01-01 0.2932 2.1593 0.2777
2000-01-02 0.4830 0.8858 0.9068
2000-01-03 2.0053 1.4074 2.6110
2000-01-04 0.3294 0.5380 1.1615
2000-01-05 0.4808 1.9892 1.1930
2000-01-06 1.4968 0.8565 1.3521
2000-01-07 0.1131 0.2541 0.3851
2000-01-08 4.3176 0.1750 0.4375
In [97]: np.asarray(df)
Out[97]:
array([[-1.2268, 0.7698, -1.2812],
[-0.7277, -0.1213, -0.0979],
[ 0.6958, 0.3417, 0.9597],
[-1.1103, -0.62 , 0.1497],
[-0.7323, 0.6877, 0.1764],
[ 0.4033, -0.155 , 0.3016],
[-2.1799, -1.3698, -0.9542],
[ 1.4627, -1.7432, -0.8266]])
点方法DataFrame实现矩阵乘法:
In [98]: df.T.dot(df)
Out[98]:
A B C
A 11.3419 -0.0598 3.0080
B -0.0598 6.5206 2.0833
C 3.0080 2.0833 4.3105
同样,点方法实现点系列产品:
In [99]: s1 = pd.Series(np.arange(5,10))
In [100]: s1.dot(s1)
Out[100]: 255
DataFrame并不打算ndarray作为替代 语义索引的地方有很大的不同从一个矩阵。
控制台显示
非常大的DataFrames将被截断在控制台显示它们。 你也可以使用 信息() 。 (我在这里读的CSV版本 棒球 数据集的 plyr R包):
In [101]: baseball = pd.read_csv('data/baseball.csv')
In [102]: print(baseball)
id player year stint ... hbp sh sf gidp
0 88641 womacto01 2006 2 ... 0.0 3.0 0.0 0.0
1 88643 schilcu01 2006 1 ... 0.0 0.0 0.0 0.0
.. ... ... ... ... ... ... ... ... ...
98 89533 aloumo01 2007 1 ... 2.0 0.0 3.0 13.0
99 89534 alomasa02 2007 1 ... 0.0 0.0 0.0 0.0
[100 rows x 23 columns]
In [103]: baseball.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 100 entries, 0 to 99
Data columns (total 23 columns):
id 100 non-null int64
player 100 non-null object
year 100 non-null int64
stint 100 non-null int64
team 100 non-null object
lg 100 non-null object
g 100 non-null int64
ab 100 non-null int64
r 100 non-null int64
h 100 non-null int64
X2b 100 non-null int64
X3b 100 non-null int64
hr 100 non-null int64
rbi 100 non-null float64
sb 100 non-null float64
cs 100 non-null float64
bb 100 non-null int64
so 100 non-null float64
ibb 100 non-null float64
hbp 100 non-null float64
sh 100 non-null float64
sf 100 non-null float64
gidp 100 non-null float64
dtypes: float64(9), int64(11), object(3)
memory usage: 18.0+ KB
然而,使用 to_string 将返回一个字符串表示的吗 DataFrame以表格形式,尽管它并不总是适合控制台宽度:
In [104]: print(baseball.iloc[-20:, :12].to_string())
id player year stint team lg g ab r h X2b X3b
80 89474 finlest01 2007 1 COL NL 43 94 9 17 3 0
81 89480 embreal01 2007 1 OAK AL 4 0 0 0 0 0
82 89481 edmonji01 2007 1 SLN NL 117 365 39 92 15 2
83 89482 easleda01 2007 1 NYN NL 76 193 24 54 6 0
84 89489 delgaca01 2007 1 NYN NL 139 538 71 139 30 0
85 89493 cormirh01 2007 1 CIN NL 6 0 0 0 0 0
86 89494 coninje01 2007 2 NYN NL 21 41 2 8 2 0
87 89495 coninje01 2007 1 CIN NL 80 215 23 57 11 1
88 89497 clemero02 2007 1 NYA AL 2 2 0 1 0 0
89 89498 claytro01 2007 2 BOS AL 8 6 1 0 0 0
90 89499 claytro01 2007 1 TOR AL 69 189 23 48 14 0
91 89501 cirilje01 2007 2 ARI NL 28 40 6 8 4 0
92 89502 cirilje01 2007 1 MIN AL 50 153 18 40 9 2
93 89521 bondsba01 2007 1 SFN NL 126 340 75 94 14 0
94 89523 biggicr01 2007 1 HOU NL 141 517 68 130 31 3
95 89525 benitar01 2007 2 FLO NL 34 0 0 0 0 0
96 89526 benitar01 2007 1 SFN NL 19 0 0 0 0 0
97 89530 ausmubr01 2007 1 HOU NL 117 349 38 82 16 3
98 89533 aloumo01 2007 1 NYN NL 87 328 51 112 19 1
99 89534 alomasa02 2007 1 NYN NL 8 22 1 3 1 0
宽DataFrames将打印多个行 默认值:
In [105]: pd.DataFrame(np.random.randn(3, 12))
Out[105]:
0 1 2 3 4 5 6 \
0 -0.345352 1.314232 0.690579 0.995761 2.396780 0.014871 3.357427
1 -2.182937 0.380396 0.084844 0.432390 1.519970 -0.493662 0.600178
2 0.206053 -0.251905 -2.213588 1.063327 1.266143 0.299368 -0.863838
7 8 9 10 11
0 -0.317441 -1.236269 0.896171 -0.487602 -0.082240
1 0.274230 0.132885 -0.023688 2.410179 1.450520
2 0.408204 -1.048089 -0.025747 -0.988387 0.094055
你可以改变多少打印一行通过设置 display.width 选择:
In [106]: pd.set_option('display.width', 40) # default is 80
In [107]: pd.DataFrame(np.random.randn(3, 12))
Out[107]:
0 1 2 \
0 1.262731 1.289997 0.082423
1 1.126203 -0.977349 1.474071
2 0.758527 1.729689 -0.964980
3 4 5 \
0 -0.055758 0.536580 -0.489682
1 -0.064034 -1.282782 0.781836
2 -0.845696 -1.340896 1.846883
6 7 8 \
0 0.369374 -0.034571 -2.484478
1 -1.071357 0.441153 2.353925
2 -1.328865 1.682706 -1.717693
9 10 11
0 -0.281461 0.030711 0.109121
1 0.583787 0.221471 -0.744471
2 0.888782 0.228440 0.901805
你可以调整的最大单个列的宽度设置 display.max_colwidth
In [108]: datafile={'filename': ['filename_01','filename_02'],
.....: 'path': ["media/user_name/storage/folder_01/filename_01",
.....: "media/user_name/storage/folder_02/filename_02"]}
.....:
In [109]: pd.set_option('display.max_colwidth',30)
In [110]: pd.DataFrame(datafile)
Out[110]:
filename \
0 filename_01
1 filename_02
path
0 media/user_name/storage/fo...
1 media/user_name/storage/fo...
In [111]: pd.set_option('display.max_colwidth',100)
In [112]: pd.DataFrame(datafile)
Out[112]:
filename \
0 filename_01
1 filename_02
path
0 media/user_name/storage/folder_01/filename_01
1 media/user_name/storage/folder_02/filename_02
你也可以禁用这个特性通过 expand_frame_repr 选择。 这将打印表在一块。
DataFrame列属性访问和IPython完成
如果DataFrame列标签是一个有效的Python变量名,列 像访问属性:
In [113]: df = pd.DataFrame({'foo1' : np.random.randn(5),
.....: 'foo2' : np.random.randn(5)})
.....:
In [114]: df
Out[114]:
foo1 foo2
0 1.171216 -0.858447
1 0.520260 0.306996
2 -1.197071 -0.028665
3 -1.066969 0.384316
4 -0.303421 1.574159
In [115]: df.foo1
Out[115]:
0 1.171216
1 0.520260
2 -1.197071
3 -1.066969
4 -0.303421
Name: foo1, dtype: float64
列也连接到 IPython 补全机制,这样他们可以tab-completed:
In [5]: df.fo<TAB>
df.foo1 df.foo2
面板 (暂时跳过)
面板有点不常用,但仍然重要的三维容器 数据。 这个词 面板数据 是 来自计量经济学和部分负责熊猫名字: 锅(el)- da(ta)- s。 3轴的名字是为了给一些语义 意义包括面板数据和描述操作,特别是, 面板数据的计量经济分析。 然而,对于严格的切片的目的 和切割DataFrame对象的集合,你会发现轴名称 稍微随意:
- 项目 :轴0,每一项对应一个DataFrame包含在里面
- major_axis :轴1, 指数 (行)的每一个 DataFrames
- minor_axis :轴2, 列 每个DataFrames的
建设小组工作大约像你期望:
与可选的3 d ndarray轴标签
In [116]: wp = pd.Panel(np.random.randn(2, 5, 4), items=['Item1', 'Item2'],
.....: major_axis=pd.date_range('1/1/2000', periods=5),
.....: minor_axis=['A', 'B', 'C', 'D'])
.....:
In [117]: wp
Out[117]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 5 (major_axis) x 4 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to D
从DataFrame dict对象
In [118]: data = {'Item1' : pd.DataFrame(np.random.randn(4, 3)),
.....: 'Item2' : pd.DataFrame(np.random.randn(4, 2))}
.....:
In [119]: pd.Panel(data)
Out[119]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 4 (major_axis) x 3 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 0 to 3
Minor_axis axis: 0 to 2
请注意,关键字的值只需要 可转换到 DataFrame 。 因此,他们可以是任何其他有效的DataFrame作为输入 /以上。
一个有用的工厂方法 Panel.from_dict 一个 字典DataFrames如上所述,以下命名参数:
| 参数 | 默认的 | 描述 |
|---|---|---|
| 相交 | 假 |
滴元素的指标不一致 |
| 东方 | 项目 |
使用 小 使用DataFrames”列小组项目 |
例如,与上面的建设:
In [120]: pd.Panel.from_dict(data, orient='minor')
Out[120]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 4 (major_axis) x 2 (minor_axis)
Items axis: 0 to 2
Major_axis axis: 0 to 3
Minor_axis axis: Item1 to Item2
东方是特别有用的混合型DataFrames。 如果你通过的东西 DataFrame对象与混合型列,所有数据会向上抛 dtype =对象 除非你通过 东方= '小' :
In [121]: df = pd.DataFrame({'a': ['foo', 'bar', 'baz'],
.....: 'b': np.random.randn(3)})
.....:
In [122]: df
Out[122]:
a b
0 foo -0.308853
1 bar -0.681087
2 baz 0.377953
In [123]: data = {'item1': df, 'item2': df}
In [124]: panel = pd.Panel.from_dict(data, orient='minor')
In [125]: panel['a']
Out[125]:
item1 item2
0 foo foo
1 bar bar
2 baz baz
In [126]: panel['b']
Out[126]:
item1 item2
0 -0.308853 -0.308853
1 -0.681087 -0.681087
2 0.377953 0.377953
In [127]: panel['b'].dtypes
Out[127]:
item1 float64
item2 float64
dtype: object
请注意
面板,比系列和DataFrame不常用, 已经有点被忽视的feature-wise。 一系列的方法和选择 在面板可用在DataFrame并不可用。
从DataFrame使用 to_panel 方法
to_panel 将一个DataFrame两级指数转换为面板。
In [128]: midx = pd.MultiIndex(levels=[['one', 'two'], ['x','y']], labels=[[1,1,0,0],[1,0,1,0]])
In [129]: df = pd.DataFrame({'A' : [1, 2, 3, 4], 'B': [5, 6, 7, 8]}, index=midx)
In [130]: df.to_panel()
Out[130]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 2 (major_axis) x 2 (minor_axis)
Items axis: A to B
Major_axis axis: one to two
Minor_axis axis: x to y
项目选择/添加/删除
类似DataFrame功能的dict系列,小组就像一个东西 DataFrames:
In [131]: wp['Item1']
Out[131]:
A B C D
2000-01-01 1.588931 0.476720 0.473424 -0.242861
2000-01-02 -0.014805 -0.284319 0.650776 -1.461665
2000-01-03 -1.137707 -0.891060 -0.693921 1.613616
2000-01-04 0.464000 0.227371 -0.496922 0.306389
2000-01-05 -2.290613 -1.134623 -1.561819 -0.260838
In [132]: wp['Item3'] = wp['Item1'] / wp['Item2']
插入和删除的API为DataFrame是一样的。 与 python DataFrame,如果项目是一个有效的标识符,你可以作为一个访问 属性和在IPython tab-complete它。
置换
一个小组可以重新使用它 转置 方法(不做 复制在默认情况下,除非数据异构):
In [133]: wp.transpose(2, 0, 1)
Out[133]:
<class 'pandas.core.panel.Panel'>
Dimensions: 4 (items) x 3 (major_axis) x 5 (minor_axis)
Items axis: A to D
Major_axis axis: Item1 to Item3
Minor_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00
索引/选择
| 操作 | 语法 | 结果 |
|---|---|---|
| 选择项 | wp(项) |
DataFrame |
| 片在major_axis标签 | wp.major_xs(val) |
DataFrame |
| 片在minor_axis标签 | wp.minor_xs(val) |
DataFrame |
例如,使用前面的例子的数据,我们能做的:
In [134]: wp['Item1']
Out[134]:
A B C D
2000-01-01 1.588931 0.476720 0.473424 -0.242861
2000-01-02 -0.014805 -0.284319 0.650776 -1.461665
2000-01-03 -1.137707 -0.891060 -0.693921 1.613616
2000-01-04 0.464000 0.227371 -0.496922 0.306389
2000-01-05 -2.290613 -1.134623 -1.561819 -0.260838
In [135]: wp.major_xs(wp.major_axis[2])
Out[135]:
Item1 Item2 Item3
A -1.137707 0.800193 -1.421791
B -0.891060 0.782098 -1.139320
C -0.693921 -1.069094 0.649074
D 1.613616 -1.099248 -1.467927
In [136]: wp.minor_axis
Out[136]: Index(['A', 'B', 'C', 'D'], dtype='object')
In [137]: wp.minor_xs('C')
Out[137]:
Item1 Item2 Item3
2000-01-01 0.473424 -0.902937 -0.524316
2000-01-02 0.650776 -1.144073 -0.568824
2000-01-03 -0.693921 -1.069094 0.649074
2000-01-04 -0.496922 0.661084 -0.751678
2000-01-05 -1.561819 -1.056652 1.478083
挤压
改变一个对象的维数的另一种方法是 挤压 1-len对象,类似 wp(“Item1”)
In [138]: wp.reindex(items=['Item1']).squeeze()
Out[138]:
A B C D
2000-01-01 1.588931 0.476720 0.473424 -0.242861
2000-01-02 -0.014805 -0.284319 0.650776 -1.461665
2000-01-03 -1.137707 -0.891060 -0.693921 1.613616
2000-01-04 0.464000 0.227371 -0.496922 0.306389
2000-01-05 -2.290613 -1.134623 -1.561819 -0.260838
In [139]: wp.reindex(items=['Item1'], minor=['B']).squeeze()
Out[139]:
2000-01-01 0.476720
2000-01-02 -0.284319
2000-01-03 -0.891060
2000-01-04 0.227371
2000-01-05 -1.134623
Freq: D, Name: B, dtype: float64
转换为DataFrame
面板可以用2 d表示形式作为分层索引 DataFrame。 看到的部分 分层索引 更多的在这。 转换一个DataFrame面板,使用 to_frame 方法:
In [140]: panel = pd.Panel(np.random.randn(3, 5, 4), items=['one', 'two', 'three'],
.....: major_axis=pd.date_range('1/1/2000', periods=5),
.....: minor_axis=['a', 'b', 'c', 'd'])
.....:
In [141]: panel.to_frame()
Out[141]:
one two three
major minor
2000-01-01 a 0.493672 1.219492 -1.290493
b -2.461467 0.062297 0.787872
c -1.553902 -0.110388 1.515707
d 2.015523 -1.184357 -0.276487
2000-01-02 a -1.833722 -0.558081 -0.223762
b 1.771740 0.077849 1.397431
c -0.670027 0.629498 1.503874
d 0.049307 -1.035260 -0.478905
2000-01-03 a -0.521493 -0.438229 -0.135950
b -3.201750 0.503703 -0.730327
c 0.792716 0.413086 -0.033277
d 0.146111 -1.139050 0.281151
2000-01-04 a 1.903247 0.660342 -1.298915
b -0.747169 0.464794 -2.819487
c -0.309038 -0.309337 -0.851985
d 0.393876 -0.649593 -1.106952
2000-01-05 a 1.861468 0.683758 -0.937731
b 0.936527 -0.643834 -1.537770
c 1.255746 0.421287 0.555759
d -2.655452 1.032814 -2.277282
反对面板
在过去的几年里,大熊猫在广度和深度,增加了新功能, 数据类型的支持,和操作例程。 因此,支持高效的索引和功能 例程 系列 , DataFrame 和 面板 导致了越来越分散, 难以理解的代码库。
的三维结构 面板 是更常见的对于许多类型的数据分析, 不是一维的 系列 或二维的 DataFrame。 未来是有道理的 熊猫只专注于这些领域。
通常,一个可以简单地使用一个MultiIndex DataFrame 轻松地处理高维数据。
此外, xarray 包是从头构建,专门为了 支持多维分析就是其中之一 面板 年代主要可变性。 这里是一个链接到xarray panel-transition文档 。
In [142]: p = tm.makePanel()
In [143]: p
Out[143]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 30 (major_axis) x 4 (minor_axis)
Items axis: ItemA to ItemC
Major_axis axis: 2000-01-03 00:00:00 to 2000-02-11 00:00:00
Minor_axis axis: A to D
转换为一个MultiIndex DataFrame
In [144]: p.to_frame()
Out[144]:
ItemA ItemB ItemC
major minor
2000-01-03 A -0.390201 -1.624062 -0.605044
B 1.562443 0.483103 0.583129
C -1.085663 0.768159 -0.273458
D 0.136235 -0.021763 -0.700648
2000-01-04 A 1.207122 -0.758514 0.878404
B 0.763264 0.061495 -0.876690
C -1.114738 0.225441 -0.335117
D 0.886313 -0.047152 -1.166607
2000-01-05 A 0.178690 -0.560859 -0.921485
B 0.162027 0.240767 -1.919354
C -0.058216 0.543294 -0.476268
D -1.350722 0.088472 -0.367236
2000-01-06 A -1.004168 -0.589005 -0.200312
B -0.902704 0.782413 -0.572707
C -0.486768 0.771931 -1.765602
D -0.886348 -0.857435 1.296674
2000-01-07 A -1.377627 -1.070678 0.522423
B 1.106010 0.628462 -1.736484
C 1.685148 -0.968145 0.578223
D -1.013316 -2.503786 0.641385
2000-01-10 A 0.499281 -1.681101 0.722511
B -0.199234 -0.880627 -1.335113
C 0.112572 -1.176383 0.242697
D 1.920906 -1.058041 -0.779432
2000-01-11 A -1.405256 0.403776 -1.702486
B 0.458265 0.777575 -1.244471
C -1.495309 -3.192716 0.208129
D -0.388231 -0.657981 0.602456
2000-01-12 A 0.162565 0.609862 -0.709535
B 0.491048 -0.779367 0.347339
... ... ... ...
2000-02-02 C -0.303961 -0.463752 -0.288962
D 0.104050 1.116086 0.506445
2000-02-03 A -2.338595 -0.581967 -0.801820
B -0.557697 -0.033731 -0.176382
C 0.625555 -0.055289 0.875359
D 0.174068 -0.443915 1.626369
2000-02-04 A -0.374279 -1.233862 -0.915751
B 0.381353 -1.108761 -1.970108
C -0.059268 -0.360853 -0.614618
D -0.439461 -0.200491 0.429518
2000-02-07 A -2.359958 -3.520876 -0.288156
B 1.337122 -0.314399 -1.044208
C 0.249698 0.728197 0.565375
D -0.741343 1.092633 0.013910
2000-02-08 A -1.157886 0.516870 -1.199945
B -1.531095 -0.860626 -0.821179
C 1.103949 1.326768 0.068184
D -0.079673 -1.675194 -0.458272
2000-02-09 A -0.551865 0.343125 -0.072869
B 1.331458 0.370397 -1.914267
C -1.087532 0.208927 0.788871
D -0.922875 0.437234 -1.531004
2000-02-10 A 1.592673 2.137827 -1.828740
B -0.571329 -1.761442 -0.826439
C 1.998044 0.292058 -0.280343
D 0.303638 0.388254 -0.500569
2000-02-11 A 1.559318 0.452429 -1.716981
B -0.026671 -0.899454 0.124808
C -0.244548 -2.019610 0.931536
D -0.917368 0.479630 0.870690
[120 rows x 3 columns]
另外,一个可以转换为一个xarray DataArray 。
In [145]: p.to_xarray()
Out[145]:
<xarray.DataArray (items: 3, major_axis: 30, minor_axis: 4)>
array([[[-0.390201, 1.562443, -1.085663, 0.136235],
[ 1.207122, 0.763264, -1.114738, 0.886313],
...,
[ 1.592673, -0.571329, 1.998044, 0.303638],
[ 1.559318, -0.026671, -0.244548, -0.917368]],
[[-1.624062, 0.483103, 0.768159, -0.021763],
[-0.758514, 0.061495, 0.225441, -0.047152],
...,
[ 2.137827, -1.761442, 0.292058, 0.388254],
[ 0.452429, -0.899454, -2.01961 , 0.47963 ]],
[[-0.605044, 0.583129, -0.273458, -0.700648],
[ 0.878404, -0.87669 , -0.335117, -1.166607],
...,
[-1.82874 , -0.826439, -0.280343, -0.500569],
[-1.716981, 0.124808, 0.931536, 0.87069 ]]])
Coordinates:
* items (items) object 'ItemA' 'ItemB' 'ItemC'
* major_axis (major_axis) datetime64[ns] 2000-01-03 2000-01-04 2000-01-05 ...
* minor_axis (minor_axis) object 'A' 'B' 'C' 'D'
你可以看到的全部文档 xarray包 。
Panel4D和PanelND(弃用)
警告
在0.19.0 Panel4D 和 PanelND 弃用和将被删除吗 未来的版本。 推荐的方式来表示这些类型的 n维的数据 xarray包 。 大熊猫提供了一个 to_xarray() 方法来自动化 这种转换。
看到 前一版本的文档 这些对象的文档。
------------------------------------------------------------------------------------
Creating a Series by passing a list of values, letting pandas create a default integer index:
In [4]: s = pd.Series([1,3,5,np.nan,6,8])
In [5]: s
Out[5]:
0 1.0
1 3.0
2 5.0
3 NaN
4 6.0
5 8.0
dtype: float64
Creating a DataFrame by passing a numpy array, with a datetime index and labeled columns:
In [6]: dates = pd.date_range('20130101', periods=6)
In [7]: dates
Out[7]:
DatetimeIndex(['2013-01-01', '2013-01-02', '2013-01-03', '2013-01-04',
'2013-01-05', '2013-01-06'],
dtype='datetime64[ns]', freq='D')
In [8]: df = pd.DataFrame(np.random.randn(6,4), index=dates, columns=list('ABCD'))
In [9]: df
Out[9]:
A B C D
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
2013-01-06 -0.673690 0.113648 -1.478427 0.524988
Creating a DataFrame by passing a dict of objects that can be converted to series-like.
In [10]: df2 = pd.DataFrame({ 'A' : 1.,
....: 'B' : pd.Timestamp('20130102'),
....: 'C' : pd.Series(1,index=list(range(4)),dtype='float32'),
....: 'D' : np.array([3] * 4,dtype='int32'),
....: 'E' : pd.Categorical(["test","train","test","train"]),
....: 'F' : 'foo' })
....:
In [11]: df2
Out[11]:
A B C D E F
0 1.0 2013-01-02 1.0 3 test foo
1 1.0 2013-01-02 1.0 3 train foo
2 1.0 2013-01-02 1.0 3 test foo
3 1.0 2013-01-02 1.0 3 train foo
Having specific dtypes
In [12]: df2.dtypes
Out[12]:
A float64
B datetime64[ns]
C float32
D int32
E category
F object
dtype: object
If you’re using IPython, tab completion for column names (as well as public attributes) is automatically enabled. Here’s a subset of the attributes that will be completed:
In [13]: df2.<TAB>
df2.A df2.bool
df2.abs df2.boxplot
df2.add df2.C
df2.add_prefix df2.clip
df2.add_suffix df2.clip_lower
df2.align df2.clip_upper
df2.all df2.columns
df2.any df2.combine
df2.append df2.combine_first
df2.apply df2.compound
df2.applymap df2.consolidate
df2.D
As you can see, the columns A, B, C, and D are automatically tab completed. E is there as well; the rest of the attributes have been truncated for brevity.
Viewing Data
See the Basics section
See the top & bottom rows of the frame
In [14]: df.head()
Out[14]:
A B C D
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
In [15]: df.tail(3)
Out[15]:
A B C D
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
2013-01-06 -0.673690 0.113648 -1.478427 0.524988
Display the index, columns, and the underlying numpy data
In [16]: df.index
Out[16]:
DatetimeIndex(['2013-01-01', '2013-01-02', '2013-01-03', '2013-01-04',
'2013-01-05', '2013-01-06'],
dtype='datetime64[ns]', freq='D')
In [17]: df.columns
Out[17]: Index(['A', 'B', 'C', 'D'], dtype='object')
In [18]: df.values
Out[18]:
array([[ 0.4691, -0.2829, -1.5091, -1.1356],
[ 1.2121, -0.1732, 0.1192, -1.0442],
[-0.8618, -2.1046, -0.4949, 1.0718],
[ 0.7216, -0.7068, -1.0396, 0.2719],
[-0.425 , 0.567 , 0.2762, -1.0874],
[-0.6737, 0.1136, -1.4784, 0.525 ]])
Describe shows a quick statistic summary of your data
In [19]: df.describe()
Out[19]:
A B C D
count 6.000000 6.000000 6.000000 6.000000
mean 0.073711 -0.431125 -0.687758 -0.233103
std 0.843157 0.922818 0.779887 0.973118
min -0.861849 -2.104569 -1.509059 -1.135632
25% -0.611510 -0.600794 -1.368714 -1.076610
50% 0.022070 -0.228039 -0.767252 -0.386188
75% 0.658444 0.041933 -0.034326 0.461706
max 1.212112 0.567020 0.276232 1.071804
Transposing your data
In [20]: df.T
Out[20]:
2013-01-01 2013-01-02 2013-01-03 2013-01-04 2013-01-05 2013-01-06
A 0.469112 1.212112 -0.861849 0.721555 -0.424972 -0.673690
B -0.282863 -0.173215 -2.104569 -0.706771 0.567020 0.113648
C -1.509059 0.119209 -0.494929 -1.039575 0.276232 -1.478427
D -1.135632 -1.044236 1.071804 0.271860 -1.087401 0.524988
Sorting by an axis
In [21]: df.sort_index(axis=1, ascending=False)
Out[21]:
D C B A
2013-01-01 -1.135632 -1.509059 -0.282863 0.469112
2013-01-02 -1.044236 0.119209 -0.173215 1.212112
2013-01-03 1.071804 -0.494929 -2.104569 -0.861849
2013-01-04 0.271860 -1.039575 -0.706771 0.721555
2013-01-05 -1.087401 0.276232 0.567020 -0.424972
2013-01-06 0.524988 -1.478427 0.113648 -0.673690
Sorting by values
In [22]: df.sort_values(by='B')
Out[22]:
A B C D
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-06 -0.673690 0.113648 -1.478427 0.524988
2013-01-05 -0.424972 0.567020 0.276232 -1.087401
Selection
Note
While standard Python / Numpy expressions for selecting and setting are intuitive and come in handy for interactive work, for production code, we recommend the optimized pandas data access methods, .at, .iat, .loc,.iloc and .ix.
See the indexing documentation Indexing and Selecting Data and MultiIndex / Advanced Indexing
Getting
Selecting a single column, which yields a Series, equivalent to df.A
In [23]: df['A']
Out[23]:
2013-01-01 0.469112
2013-01-02 1.212112
2013-01-03 -0.861849
2013-01-04 0.721555
2013-01-05 -0.424972
2013-01-06 -0.673690
Freq: D, Name: A, dtype: float64
Selecting via [], which slices the rows.
In [24]: df[0:3]
Out[24]:
A B C D
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
In [25]: df['20130102':'20130104']
Out[25]:
A B C D
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
Selection by Label
See more in Selection by Label
For getting a cross section using a label
In [26]: df.loc[dates[0]]
Out[26]:
A 0.469112
B -0.282863
C -1.509059
D -1.135632
Name: 2013-01-01 00:00:00, dtype: float64
Selecting on a multi-axis by label
In [27]: df.loc[:,['A','B']]
Out[27]:
A B
2013-01-01 0.469112 -0.282863
2013-01-02 1.212112 -0.173215
2013-01-03 -0.861849 -2.104569
2013-01-04 0.721555 -0.706771
2013-01-05 -0.424972 0.567020
2013-01-06 -0.673690 0.113648
Showing label slicing, both endpoints are included
In [28]: df.loc['20130102':'20130104',['A','B']]
Out[28]:
A B
2013-01-02 1.212112 -0.173215
2013-01-03 -0.861849 -2.104569
2013-01-04 0.721555 -0.706771
Reduction in the dimensions of the returned object
In [29]: df.loc['20130102',['A','B']]
Out[29]:
A 1.212112
B -0.173215
Name: 2013-01-02 00:00:00, dtype: float64
For getting a scalar value
In [30]: df.loc[dates[0],'A']
Out[30]: 0.46911229990718628
For getting fast access to a scalar (equiv to the prior method)
In [31]: df.at[dates[0],'A']
Out[31]: 0.46911229990718628
Selection by Position
See more in Selection by Position
Select via the position of the passed integers
In [32]: df.iloc[3]
Out[32]:
A 0.721555
B -0.706771
C -1.039575
D 0.271860
Name: 2013-01-04 00:00:00, dtype: float64
By integer slices, acting similar to numpy/python
In [33]: df.iloc[3:5,0:2]
Out[33]:
A B
2013-01-04 0.721555 -0.706771
2013-01-05 -0.424972 0.567020
By lists of integer position locations, similar to the numpy/python style
In [34]: df.iloc[[1,2,4],[0,2]]
Out[34]:
A C
2013-01-02 1.212112 0.119209
2013-01-03 -0.861849 -0.494929
2013-01-05 -0.424972 0.276232
For slicing rows explicitly
In [35]: df.iloc[1:3,:]
Out[35]:
A B C D
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804
For slicing columns explicitly
In [36]: df.iloc[:,1:3]
Out[36]:
B C
2013-01-01 -0.282863 -1.509059
2013-01-02 -0.173215 0.119209
2013-01-03 -2.104569 -0.494929
2013-01-04 -0.706771 -1.039575
2013-01-05 0.567020 0.276232
2013-01-06 0.113648 -1.478427
For getting a value explicitly
In [37]: df.iloc[1,1]
Out[37]: -0.17321464905330858
For getting fast access to a scalar (equiv to the prior method)
In [38]: df.iat[1,1]
Out[38]: -0.17321464905330858
Boolean Indexing
Using a single column’s values to select data.
In [39]: df[df.A > 0]
Out[39]:
A B C D
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632
2013-01-02 1.212112 -0.173215 0.119209 -1.044236
2013-01-04 0.721555 -0.706771 -1.039575 0.271860
Selecting values from a DataFrame where a boolean condition is met.
In [40]: df[df > 0]
Out[40]:
A B C D
2013-01-01 0.469112 NaN NaN NaN
2013-01-02 1.212112 NaN 0.119209 NaN
2013-01-03 NaN NaN NaN 1.071804
2013-01-04 0.721555 NaN NaN 0.271860
2013-01-05 NaN 0.567020 0.276232 NaN
2013-01-06 NaN 0.113648 NaN 0.524988
Using the isin() method for filtering:
In [41]: df2 = df.copy()
In [42]: df2['E'] = ['one', 'one','two','three','four','three']
In [43]: df2
Out[43]:
A B C D E
2013-01-01 0.469112 -0.282863 -1.509059 -1.135632 one
2013-01-02 1.212112 -0.173215 0.119209 -1.044236 one
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804 two
2013-01-04 0.721555 -0.706771 -1.039575 0.271860 three
2013-01-05 -0.424972 0.567020 0.276232 -1.087401 four
2013-01-06 -0.673690 0.113648 -1.478427 0.524988 three
In [44]: df2[df2['E'].isin(['two','four'])]
Out[44]:
A B C D E
2013-01-03 -0.861849 -2.104569 -0.494929 1.071804 two
2013-01-05 -0.424972 0.567020 0.276232 -1.087401 four
Setting
Setting a new column automatically aligns the data by the indexes
In [45]: s1 = pd.Series([1,2,3,4,5,6], index=pd.date_range('20130102', periods=6))
In [46]: s1
Out[46]:
2013-01-02 1
2013-01-03 2
2013-01-04 3
2013-01-05 4
2013-01-06 5
2013-01-07 6
Freq: D, dtype: int64
In [47]: df['F'] = s1
Setting values by label
In [48]: df.at[dates[0],'A'] = 0
Setting values by position
In [49]: df.iat[0,1] = 0
Setting by assigning with a numpy array
In [50]: df.loc[:,'D'] = np.array([5] * len(df))
The result of the prior setting operations
In [51]: df
Out[51]:
A B C D F
2013-01-01 0.000000 0.000000 -1.509059 5 NaN
2013-01-02 1.212112 -0.173215 0.119209 5 1.0
2013-01-03 -0.861849 -2.104569 -0.494929 5 2.0
2013-01-04 0.721555 -0.706771 -1.039575 5 3.0
2013-01-05 -0.424972 0.567020 0.276232 5 4.0
2013-01-06 -0.673690 0.113648 -1.478427 5 5.0
A where operation with setting.
In [52]: df2 = df.copy()
In [53]: df2[df2 > 0] = -df2
In [54]: df2
Out[54]:
A B C D F
2013-01-01 0.000000 0.000000 -1.509059 -5 NaN
2013-01-02 -1.212112 -0.173215 -0.119209 -5 -1.0
2013-01-03 -0.861849 -2.104569 -0.494929 -5 -2.0
2013-01-04 -0.721555 -0.706771 -1.039575 -5 -3.0
2013-01-05 -0.424972 -0.567020 -0.276232 -5 -4.0
2013-01-06 -0.673690 -0.113648 -1.478427 -5 -5.0
Missing Data
pandas primarily uses the value np.nan to represent missing data. It is by default not included in computations. See the Missing Data section
Reindexing allows you to change/add/delete the index on a specified axis. This returns a copy of the data.
In [55]: df1 = df.reindex(index=dates[0:4], columns=list(df.columns) + ['E'])
In [56]: df1.loc[dates[0]:dates[1],'E'] = 1
In [57]: df1
Out[57]:
A B C D F E
2013-01-01 0.000000 0.000000 -1.509059 5 NaN 1.0
2013-01-02 1.212112 -0.173215 0.119209 5 1.0 1.0
2013-01-03 -0.861849 -2.104569 -0.494929 5 2.0 NaN
2013-01-04 0.721555 -0.706771 -1.039575 5 3.0 NaN
To drop any rows that have missing data.
In [58]: df1.dropna(how='any')
Out[58]:
A B C D F E
2013-01-02 1.212112 -0.173215 0.119209 5 1.0 1.0
Filling missing data
In [59]: df1.fillna(value=5)
Out[59]:
A B C D F E
2013-01-01 0.000000 0.000000 -1.509059 5 5.0 1.0
2013-01-02 1.212112 -0.173215 0.119209 5 1.0 1.0
2013-01-03 -0.861849 -2.104569 -0.494929 5 2.0 5.0
2013-01-04 0.721555 -0.706771 -1.039575 5 3.0 5.0
To get the boolean mask where values are nan
In [60]: pd.isna(df1)
Out[60]:
A B C D F E
2013-01-01 False False False False True False
2013-01-02 False False False False False False
2013-01-03 False False False False False True
2013-01-04 False False False False False True
Operations
See the Basic section on Binary Ops
Stats
Operations in general exclude missing data.
Performing a descriptive statistic
In [61]: df.mean()
Out[61]:
A -0.004474
B -0.383981
C -0.687758
D 5.000000
F 3.000000
dtype: float64
Same operation on the other axis
In [62]: df.mean(1)
Out[62]:
2013-01-01 0.872735
2013-01-02 1.431621
2013-01-03 0.707731
2013-01-04 1.395042
2013-01-05 1.883656
2013-01-06 1.592306
Freq: D, dtype: float64
Operating with objects that have different dimensionality and need alignment. In addition, pandas automatically broadcasts along the specified dimension.
In [63]: s = pd.Series([1,3,5,np.nan,6,8], index=dates).shift(2)
In [64]: s
Out[64]:
2013-01-01 NaN
2013-01-02 NaN
2013-01-03 1.0
2013-01-04 3.0
2013-01-05 5.0
2013-01-06 NaN
Freq: D, dtype: float64
In [65]: df.sub(s, axis='index')
Out[65]:
A B C D F
2013-01-01 NaN NaN NaN NaN NaN
2013-01-02 NaN NaN NaN NaN NaN
2013-01-03 -1.861849 -3.104569 -1.494929 4.0 1.0
2013-01-04 -2.278445 -3.706771 -4.039575 2.0 0.0
2013-01-05 -5.424972 -4.432980 -4.723768 0.0 -1.0
2013-01-06 NaN NaN NaN NaN NaN
Apply
Applying functions to the data
In [66]: df.apply(np.cumsum)
Out[66]:
A B C D F
2013-01-01 0.000000 0.000000 -1.509059 5 NaN
2013-01-02 1.212112 -0.173215 -1.389850 10 1.0
2013-01-03 0.350263 -2.277784 -1.884779 15 3.0
2013-01-04 1.071818 -2.984555 -2.924354 20 6.0
2013-01-05 0.646846 -2.417535 -2.648122 25 10.0
2013-01-06 -0.026844 -2.303886 -4.126549 30 15.0
In [67]: df.apply(lambda x: x.max() - x.min())
Out[67]:
A 2.073961
B 2.671590
C 1.785291
D 0.000000
F 4.000000
dtype: float64
Histogramming
See more at Histogramming and Discretization
In [68]: s = pd.Series(np.random.randint(0, 7, size=10))
In [69]: s
Out[69]:
0 4
1 2
2 1
3 2
4 6
5 4
6 4
7 6
8 4
9 4
dtype: int64
In [70]: s.value_counts()
Out[70]:
4 5
6 2
2 2
1 1
dtype: int64
String Methods
Series is equipped with a set of string processing methods in the str attribute that make it easy to operate on each element of the array, as in the code snippet below. Note that pattern-matching in str generally uses regular expressions by default (and in some cases always uses them). See more at Vectorized String Methods.
In [71]: s = pd.Series(['A', 'B', 'C', 'Aaba', 'Baca', np.nan, 'CABA', 'dog', 'cat'])
In [72]: s.str.lower()
Out[72]:
0 a
1 b
2 c
3 aaba
4 baca
5 NaN
6 caba
7 dog
8 cat
dtype: object
Merge
Concat
pandas provides various facilities for easily combining together Series, DataFrame, and Panel objects with various kinds of set logic for the indexes and relational algebra functionality in the case of join / merge-type operations.
See the Merging section
Concatenating pandas objects together with concat():
In [73]: df = pd.DataFrame(np.random.randn(10, 4))
In [74]: df
Out[74]:
0 1 2 3
0 -0.548702 1.467327 -1.015962 -0.483075
1 1.637550 -1.217659 -0.291519 -1.745505
2 -0.263952 0.991460 -0.919069 0.266046
3 -0.709661 1.669052 1.037882 -1.705775
4 -0.919854 -0.042379 1.247642 -0.009920
5 0.290213 0.495767 0.362949 1.548106
6 -1.131345 -0.089329 0.337863 -0.945867
7 -0.932132 1.956030 0.017587 -0.016692
8 -0.575247 0.254161 -1.143704 0.215897
9 1.193555 -0.077118 -0.408530 -0.862495
# break it into pieces
In [75]: pieces = [df[:3], df[3:7], df[7:]]
In [76]: pd.concat(pieces)
Out[76]:
0 1 2 3
0 -0.548702 1.467327 -1.015962 -0.483075
1 1.637550 -1.217659 -0.291519 -1.745505
2 -0.263952 0.991460 -0.919069 0.266046
3 -0.709661 1.669052 1.037882 -1.705775
4 -0.919854 -0.042379 1.247642 -0.009920
5 0.290213 0.495767 0.362949 1.548106
6 -1.131345 -0.089329 0.337863 -0.945867
7 -0.932132 1.956030 0.017587 -0.016692
8 -0.575247 0.254161 -1.143704 0.215897
9 1.193555 -0.077118 -0.408530 -0.862495
Join
SQL style merges. See the Database style joining
In [77]: left = pd.DataFrame({'key': ['foo', 'foo'], 'lval': [1, 2]})
In [78]: right = pd.DataFrame({'key': ['foo', 'foo'], 'rval': [4, 5]})
In [79]: left
Out[79]:
key lval
0 foo 1
1 foo 2
In [80]: right
Out[80]:
key rval
0 foo 4
1 foo 5
In [81]: pd.merge(left, right, on='key')
Out[81]:
key lval rval
0 foo 1 4
1 foo 1 5
2 foo 2 4
3 foo 2 5
Another example that can be given is:
In [82]: left = pd.DataFrame({'key': ['foo', 'bar'], 'lval': [1, 2]})
In [83]: right = pd.DataFrame({'key': ['foo', 'bar'], 'rval': [4, 5]})
In [84]: left
Out[84]:
key lval
0 foo 1
1 bar 2
In [85]: right
Out[85]:
key rval
0 foo 4
1 bar 5
In [86]: pd.merge(left, right, on='key')
Out[86]:
key lval rval
0 foo 1 4
1 bar 2 5
Append
Append rows to a dataframe. See the Appending
In [87]: df = pd.DataFrame(np.random.randn(8, 4), columns=['A','B','C','D'])
In [88]: df
Out[88]:
A B C D
0 1.346061 1.511763 1.627081 -0.990582
1 -0.441652 1.211526 0.268520 0.024580
2 -1.577585 0.396823 -0.105381 -0.532532
3 1.453749 1.208843 -0.080952 -0.264610
4 -0.727965 -0.589346 0.339969 -0.693205
5 -0.339355 0.593616 0.884345 1.591431
6 0.141809 0.220390 0.435589 0.192451
7 -0.096701 0.803351 1.715071 -0.708758
In [89]: s = df.iloc[3]
In [90]: df.append(s, ignore_index=True)
Out[90]:
A B C D
0 1.346061 1.511763 1.627081 -0.990582
1 -0.441652 1.211526 0.268520 0.024580
2 -1.577585 0.396823 -0.105381 -0.532532
3 1.453749 1.208843 -0.080952 -0.264610
4 -0.727965 -0.589346 0.339969 -0.693205
5 -0.339355 0.593616 0.884345 1.591431
6 0.141809 0.220390 0.435589 0.192451
7 -0.096701 0.803351 1.715071 -0.708758
8 1.453749 1.208843 -0.080952 -0.264610
Grouping
By “group by” we are referring to a process involving one or more of the following steps
- Splitting the data into groups based on some criteria
- Applying a function to each group independently
- Combining the results into a data structure
See the Grouping section
In [91]: df = pd.DataFrame({'A' : ['foo', 'bar', 'foo', 'bar',
....: 'foo', 'bar', 'foo', 'foo'],
....: 'B' : ['one', 'one', 'two', 'three',
....: 'two', 'two', 'one', 'three'],
....: 'C' : np.random.randn(8),
....: 'D' : np.random.randn(8)})
....:
In [92]: df
Out[92]:
A B C D
0 foo one -1.202872 -0.055224
1 bar one -1.814470 2.395985
2 foo two 1.018601 1.552825
3 bar three -0.595447 0.166599
4 foo two 1.395433 0.047609
5 bar two -0.392670 -0.136473
6 foo one 0.007207 -0.561757
7 foo three 1.928123 -1.623033
Grouping and then applying a function sum to the resulting groups.
In [93]: df.groupby('A').sum()
Out[93]:
C D
A
bar -2.802588 2.42611
foo 3.146492 -0.63958
Grouping by multiple columns forms a hierarchical index, which we then apply the function.
In [94]: df.groupby(['A','B']).sum()
Out[94]:
C D
A B
bar one -1.814470 2.395985
three -0.595447 0.166599
two -0.392670 -0.136473
foo one -1.195665 -0.616981
three 1.928123 -1.623033
two 2.414034 1.600434
Reshaping
See the sections on Hierarchical Indexing and Reshaping.
Stack
In [95]: tuples = list(zip(*[['bar', 'bar', 'baz', 'baz',
....: 'foo', 'foo', 'qux', 'qux'],
....: ['one', 'two', 'one', 'two',
....: 'one', 'two', 'one', 'two']]))
....:
In [96]: index = pd.MultiIndex.from_tuples(tuples, names=['first', 'second'])
In [97]: df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=['A', 'B'])
In [98]: df2 = df[:4]
In [99]: df2
Out[99]:
A B
first second
bar one 0.029399 -0.542108
two 0.282696 -0.087302
baz one -1.575170 1.771208
two 0.816482 1.100230
The stack() method “compresses” a level in the DataFrame’s columns.
In [100]: stacked = df2.stack()
In [101]: stacked
Out[101]:
first second
bar one A 0.029399
B -0.542108
two A 0.282696
B -0.087302
baz one A -1.575170
B 1.771208
two A 0.816482
B 1.100230
dtype: float64
With a “stacked” DataFrame or Series (having a MultiIndex as the index), the inverse operation of stack() isunstack(), which by default unstacks the last levelIn [102]: stacked.unstack()Out[102]:
A B
first second
bar one 0.029399 -0.542108
two 0.282696 -0.087302
baz one -1.575170 1.771208
two 0.816482 1.100230
In [103]: stacked.unstack(1)
Out[103]:
second one two
first
bar A 0.029399 0.282696
B -0.542108 -0.087302
baz A -1.575170 0.816482
B 1.771208 1.100230
In [104]: stacked.unstack(0)
Out[104]:
first bar baz
second
one A 0.029399 -1.575170
B -0.542108 1.771208
two A 0.282696 0.816482
B -0.087302 1.100230
------------------------------------------------------------------------------------------------------------------------------
此处以下,以后再看。
Pivot Tables
See the section on Pivot Tables.
In [105]: df = pd.DataFrame({'A' : ['one', 'one', 'two', 'three'] * 3,
.....: 'B' : ['A', 'B', 'C'] * 4,
.....: 'C' : ['foo', 'foo', 'foo', 'bar', 'bar', 'bar'] * 2,
.....: 'D' : np.random.randn(12),
.....: 'E' : np.random.randn(12)})
.....:
In [106]: df
Out[106]:
A B C D E
0 one A foo 1.418757 -0.179666
1 one B foo -1.879024 1.291836
2 two C foo 0.536826 -0.009614
3 three A bar 1.006160 0.392149
4 one B bar -0.029716 0.264599
5 one C bar -1.146178 -0.057409
6 two A foo 0.100900 -1.425638
7 three B foo -1.035018 1.024098
8 one C foo 0.314665 -0.106062
9 one A bar -0.773723 1.824375
10 two B bar -1.170653 0.595974
11 three C bar 0.648740 1.167115
We can produce pivot tables from this data very easily:
In [107]: pd.pivot_table(df, values='D', index=['A', 'B'], columns=['C'])
Out[107]:
C bar foo
A B
one A -0.773723 1.418757
B -0.029716 -1.879024
C -1.146178 0.314665
three A 1.006160 NaN
B NaN -1.035018
C 0.648740 NaN
two A NaN 0.100900
B -1.170653 NaN
C NaN 0.536826
Time Series
pandas has simple, powerful, and efficient functionality for performing resampling operations during frequency conversion (e.g., converting secondly data into 5-minutely data). This is extremely common in, but not limited to, financial applications. See the Time Series section
In [108]: rng = pd.date_range('1/1/2012', periods=100, freq='S')
In [109]: ts = pd.Series(np.random.randint(0, 500, len(rng)), index=rng)
In [110]: ts.resample('5Min').sum()
Out[110]:
2012-01-01 25083
Freq: 5T, dtype: int64
Time zone representation
In [111]: rng = pd.date_range('3/6/2012 00:00', periods=5, freq='D')
In [112]: ts = pd.Series(np.random.randn(len(rng)), rng)
In [113]: ts
Out[113]:
2012-03-06 0.464000
2012-03-07 0.227371
2012-03-08 -0.496922
2012-03-09 0.306389
2012-03-10 -2.290613
Freq: D, dtype: float64
In [114]: ts_utc = ts.tz_localize('UTC')
In [115]: ts_utc
Out[115]:
2012-03-06 00:00:00+00:00 0.464000
2012-03-07 00:00:00+00:00 0.227371
2012-03-08 00:00:00+00:00 -0.496922
2012-03-09 00:00:00+00:00 0.306389
2012-03-10 00:00:00+00:00 -2.290613
Freq: D, dtype: float64
Convert to another time zone
In [116]: ts_utc.tz_convert('US/Eastern')
Out[116]:
2012-03-05 19:00:00-05:00 0.464000
2012-03-06 19:00:00-05:00 0.227371
2012-03-07 19:00:00-05:00 -0.496922
2012-03-08 19:00:00-05:00 0.306389
2012-03-09 19:00:00-05:00 -2.290613
Freq: D, dtype: float64
Converting between time span representations
In [117]: rng = pd.date_range('1/1/2012', periods=5, freq='M')
In [118]: ts = pd.Series(np.random.randn(len(rng)), index=rng)
In [119]: ts
Out[119]:
2012-01-31 -1.134623
2012-02-29 -1.561819
2012-03-31 -0.260838
2012-04-30 0.281957
2012-05-31 1.523962
Freq: M, dtype: float64
In [120]: ps = ts.to_period()
In [121]: ps
Out[121]:
2012-01 -1.134623
2012-02 -1.561819
2012-03 -0.260838
2012-04 0.281957
2012-05 1.523962
Freq: M, dtype: float64
In [122]: ps.to_timestamp()
Out[122]:
2012-01-01 -1.134623
2012-02-01 -1.561819
2012-03-01 -0.260838
2012-04-01 0.281957
2012-05-01 1.523962
Freq: MS, dtype: float64
Converting between period and timestamp enables some convenient arithmetic functions to be used. In the following example, we convert a quarterly frequency with year ending in November to 9am of the end of the month following the quarter end:
In [123]: prng = pd.period_range('1990Q1', '2000Q4', freq='Q-NOV')
In [124]: ts = pd.Series(np.random.randn(len(prng)), prng)
In [125]: ts.index = (prng.asfreq('M', 'e') + 1).asfreq('H', 's') + 9
In [126]: ts.head()
Out[126]:
1990-03-01 09:00 -0.902937
1990-06-01 09:00 0.068159
1990-09-01 09:00 -0.057873
1990-12-01 09:00 -0.368204
1991-03-01 09:00 -1.144073
Freq: H, dtype: float64
Categoricals
pandas can include categorical data in a DataFrame. For full docs, see the categorical introduction and the API documentation.
In [127]: df = pd.DataFrame({"id":[1,2,3,4,5,6], "raw_grade":['a', 'b', 'b', 'a', 'a', 'e']})
Convert the raw grades to a categorical data type.
In [128]: df["grade"] = df["raw_grade"].astype("category")
In [129]: df["grade"]
Out[129]:
0 a
1 b
2 b
3 a
4 a
5 e
Name: grade, dtype: category
Categories (3, object): [a, b, e]
Rename the categories to more meaningful names (assigning to Series.cat.categories is inplace!)
In [130]: df["grade"].cat.categories = ["very good", "good", "very bad"]
Reorder the categories and simultaneously add the missing categories (methods under Series .cat return a new Series per default).
In [131]: df["grade"] = df["grade"].cat.set_categories(["very bad", "bad", "medium", "good", "very good"])
In [132]: df["grade"]
Out[132]:
0 very good
1 good
2 good
3 very good
4 very good
5 very bad
Name: grade, dtype: category
Categories (5, object): [very bad, bad, medium, good, very good]
Sorting is per order in the categories, not lexical order.
In [133]: df.sort_values(by="grade")
Out[133]:
id raw_grade grade
5 6 e very bad
1 2 b good
2 3 b good
0 1 a very good
3 4 a very good
4 5 a very good
Grouping by a categorical column shows also empty categories.
In [134]: df.groupby("grade").size()
Out[134]:
grade
very bad 1
bad 0
medium 0
good 2
very good 3
dtype: int64
Plotting
Plotting docs.
In [135]: ts = pd.Series(np.random.randn(1000), index=pd.date_range('1/1/2000', periods=1000))
In [136]: ts = ts.cumsum()
In [137]: ts.plot()
Out[137]: <matplotlib.axes._subplots.AxesSubplot at 0x1122ad630>
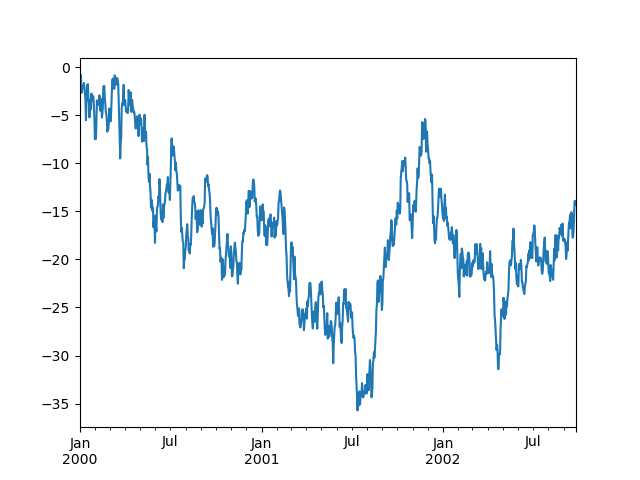
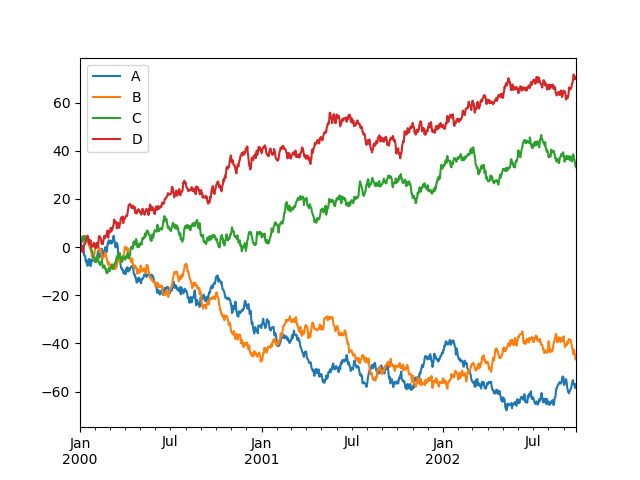
On DataFrame, plot() is a convenience to plot all of the columns with labels:
In [138]: df = pd.DataFrame(np.random.randn(1000, 4), index=ts.index,
.....: columns=['A', 'B', 'C', 'D'])
.....:
In [139]: df = df.cumsum()
In [140]: plt.figure(); df.plot(); plt.legend(loc='best')
Out[140]: <matplotlib.legend.Legend at 0x115033cf8>
Getting Data In/Out
CSV
In [141]: df.to_csv('foo.csv')
In [142]: pd.read_csv('foo.csv')
Out[142]:
Unnamed: 0 A B C D
0 2000-01-01 0.266457 -0.399641 -0.219582 1.186860
1 2000-01-02 -1.170732 -0.345873 1.653061 -0.282953
2 2000-01-03 -1.734933 0.530468 2.060811 -0.515536
3 2000-01-04 -1.555121 1.452620 0.239859 -1.156896
4 2000-01-05 0.578117 0.511371 0.103552 -2.428202
5 2000-01-06 0.478344 0.449933 -0.741620 -1.962409
6 2000-01-07 1.235339 -0.091757 -1.543861 -1.084753
.. ... ... ... ... ...
993 2002-09-20 -10.628548 -9.153563 -7.883146 28.313940
994 2002-09-21 -10.390377 -8.727491 -6.399645 30.914107
995 2002-09-22 -8.985362 -8.485624 -4.669462 31.367740
996 2002-09-23 -9.558560 -8.781216 -4.499815 30.518439
997 2002-09-24 -9.902058 -9.340490 -4.386639 30.105593
998 2002-09-25 -10.216020 -9.480682 -3.933802 29.758560
999 2002-09-26 -11.856774 -10.671012 -3.216025 29.369368
[1000 rows x 5 columns]
HDF5
Reading and writing to HDFStores
Writing to a HDF5 Store
In [143]: df.to_hdf('foo.h5','df')
Reading from a HDF5 Store
In [144]: pd.read_hdf('foo.h5','df')
Out[144]:
A B C D
2000-01-01 0.266457 -0.399641 -0.219582 1.186860
2000-01-02 -1.170732 -0.345873 1.653061 -0.282953
2000-01-03 -1.734933 0.530468 2.060811 -0.515536
2000-01-04 -1.555121 1.452620 0.239859 -1.156896
2000-01-05 0.578117 0.511371 0.103552 -2.428202
2000-01-06 0.478344 0.449933 -0.741620 -1.962409
2000-01-07 1.235339 -0.091757 -1.543861 -1.084753
... ... ... ... ...
2002-09-20 -10.628548 -9.153563 -7.883146 28.313940
2002-09-21 -10.390377 -8.727491 -6.399645 30.914107
2002-09-22 -8.985362 -8.485624 -4.669462 31.367740
2002-09-23 -9.558560 -8.781216 -4.499815 30.518439
2002-09-24 -9.902058 -9.340490 -4.386639 30.105593
2002-09-25 -10.216020 -9.480682 -3.933802 29.758560
2002-09-26 -11.856774 -10.671012 -3.216025 29.369368
[1000 rows x 4 columns]
Excel
Reading and writing to MS Excel
Writing to an excel file
In [145]: df.to_excel('foo.xlsx', sheet_name='Sheet1')
Reading from an excel file
In [146]: pd.read_excel('foo.xlsx', 'Sheet1', index_col=None, na_values=['NA'])
Out[146]:
A B C D
2000-01-01 0.266457 -0.399641 -0.219582 1.186860
2000-01-02 -1.170732 -0.345873 1.653061 -0.282953
2000-01-03 -1.734933 0.530468 2.060811 -0.515536
2000-01-04 -1.555121 1.452620 0.239859 -1.156896
2000-01-05 0.578117 0.511371 0.103552 -2.428202
2000-01-06 0.478344 0.449933 -0.741620 -1.962409
2000-01-07 1.235339 -0.091757 -1.543861 -1.084753
... ... ... ... ...
2002-09-20 -10.628548 -9.153563 -7.883146 28.313940
2002-09-21 -10.390377 -8.727491 -6.399645 30.914107
2002-09-22 -8.985362 -8.485624 -4.669462 31.367740
2002-09-23 -9.558560 -8.781216 -4.499815 30.518439
2002-09-24 -9.902058 -9.340490 -4.386639 30.105593
2002-09-25 -10.216020 -9.480682 -3.933802 29.758560
2002-09-26 -11.856774 -10.671012 -3.216025 29.369368
[1000 rows x 4 columns]
Gotchas
If you are trying an operation and you see an exception like:
>>> if pd.Series([False, True, False]):
print("I was true")
Traceback
...
ValueError: The truth value of an array is ambiguous. Use a.empty, a.any() or a.all().
See Comparisons for an explanation and what to do.
See Gotchas as well.







 浙公网安备 33010602011771号
浙公网安备 33010602011771号