EM and GMM(Code)
In EM and GMM(Theory), I have introduced the theory of em algorithm for gmm. Now lets practice it in matlab!
1. Generate 1000 pieces of random 2-dimention data which obey 5 gaussian distribution.
function X = GenerateData
Sigma = [1, 0; 0, 1];
mu1 = [1, -1];
x1 = mvnrnd(mu1, Sigma, 200);
mu2 = [5.5, -4.5];
x2 = mvnrnd(mu2, Sigma, 200);
mu3 = [1, 4];
x3 = mvnrnd(mu3, Sigma, 200);
mu4 = [6, 4.5];
x4 = mvnrnd(mu4, Sigma, 200);
mu5 = [9, 0.0];
x5 = mvnrnd(mu5, Sigma, 200);
% obtain the 1000 data points to be clustered
X = [x1; x2; x3; x4; x5];
end
2. Complete em algorithm.
function [Mu, Sigma, Pi, r_nk] = EmForGmm(Data, classNum, initVal)
% Data : Matrix(n * d), n is the quantity of the data and d is the data
% dimention
% classNum : Scale
% initVal : Cell(3 * 1), initial value for Mu, Sigma and Pi
% cell 1: Mu
% cell 2: Sigma
% cell 3: Pi
[sampleNum, sampleDim] = size(Data);
indexPoint = zeros(sampleNum, 1);
while(1)
for n = 1 : sampleNum
x = Data(n, :);
px_nk_sumk = 0;
for k = 1 : classNum
Sigma_k = initVal{2}(:,:,k);
Mu_k = initVal{1}(k,:);
Pi_k = initVal{3}(k);
px(n,k) = (1/(2*pi^(sampleDim/2)*det(Sigma_k)^(0.5))) ...
* exp(-0.5 * (x - Mu_k)*inv(Sigma_k)*(x - Mu_k)');
px_nk_sumk = px_nk_sumk + Pi_k * px(n, k);
end
for k = 1 : classNum
Sigma_k = initVal{2}(:,:,k);
Mu_k = initVal{1}(k,:);
Pi_k = initVal{3}(k);
r(n, k) = Pi_k * px(n, k) / px_nk_sumk;
end
end
Nk = sum(r)';
newMuK = r' * Data;
Nkk = repmat(Nk,1,2);
newMuK = newMuK ./ Nkk;
for i = 1 : classNum
nk = Nk(i);
MuT = repmat(newMuK(i,:),sampleNum,1);
xT = Data - MuT;
rT = r(:,i);
rT = repmat(rT,1,2);
newSigma(:,:,i) = xT' * (xT .* rT) / nk;
end
newPiK = Nk / sampleNum;
indexPointT = indexPoint;
[aa,indexPoint] = max(r,[],2);
j1 = sum(sum(abs(newMuK - initVal{1}))) < 1e-6;
j2 = sum(sum(sum(abs(newSigma - initVal{2})))) < 1e-6;
j3 = sum(abs(newPiK - initVal{3})) < 1e-6;
clf;
if (j1 && j2 && j3)
for i = 1:sampleNum
if (indexPoint(i)==1)
plot(Data(i,1), Data(i,2), 'r.')
end
if (indexPoint(i)==2)
plot(Data(i,1), Data(i,2), 'b.')
end
if (indexPoint(i)==3)
plot(Data(i,1), Data(i,2), 'k.')
end
if (indexPoint(i)==4)
plot(Data(i,1), Data(i,2), 'g.')
end
if (indexPoint(i)==5)
plot(Data(i,1), Data(i,2), 'm.')
end
hold on;
end
break;
else
initVal{1} = newMuK;
initVal{2} = newSigma;
initVal{3} = newPiK;
end
end
Mu = newMuK;
Sigma = newSigma;
Pi = newPiK;
r_nk = r;
end
3. Complete main function.
clear,clc,clf
Data = GenerateData;
classNum = 5;
[sampleNum, sampleDia] = size(Data);
%% Initial value
% indexNum = floor(1 + (sampleNum - 1) * rand(1,classNum));
indexNum = [50,300,500,700,900];
initMu = Data(indexNum,:);
initSigmaT = [1 0.2;0.2 1];
initSigma = zeros(2,2,classNum);
for i = 1 : classNum
initSigma(:,:,i) = initSigmaT;
initPi(i,1) = 1 / classNum;
end
initVal = cell(3,1);
initVal{1} = initMu;
initVal{2} = initSigma;
initVal{3} = initPi;
%% EM algorithm
[Mu, Sigma, Pi, r_nk] = EmForGmm(Data, classNum, initVal);
4. Result.
The cluster result can be show as figure 3.
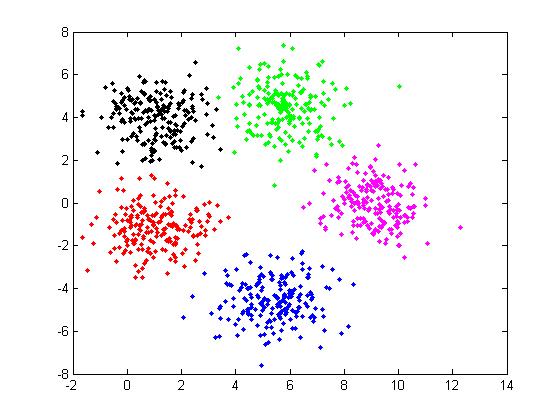
Figure 3
The probality distribution function can be writen as:
\[ p(\mathbf{x}) = \sum_{k=1}^{K}\pi_kp(\mathbf{x}|\mu_k\Sigma_k) \]
where,
$\mu_1 = (1.028, -1.158) $, $\mu_2 = (5.423, -4.538) $, $\mu_3 = (1.036, 3.975) $, $\mu_4 = (5.835, 4.474) $, $\mu_5 = (9.074, -0.063) $
Notice that, when generate the data:
$\mu_1 = (1, -1) $, $\mu_2 = (5.5, -4.5) $, $\mu_3 = (1, 4) $, $\mu_4 = (6, 4.5) $, $\mu_5 = (9, 0) $)
\[
\Sigma_1 = \left(
\begin{array}{cc}
1.0873& 0.0376\\
0.0376& 0.8850
\end{array}
\right),
\Sigma_2 = \left(
\begin{array}{cc}
1.1426& 0.0509\\
0.0509& 0.9192
\end{array}
\right),
\Sigma_3 = \left(
\begin{array}{cc}
0.9752& -0.0712\\
-0.0712& 0.9871
\end{array}
\right),
\Sigma_4 = \left(
\begin{array}{cc}
1.0111& -0.0782\\
-0.0782& 1.2034
\end{array}
\right),
\Sigma_5 = \left(
\begin{array}{cc}
0.8665& -0.1527\\
-0.1527& 0.9352
\end{array}
\right)
\]
Notice that, when generate the data:
\[\Sigma = \left(
\begin{array}{cc}
1& 0\\
0& 1
\end{array}
\right)
\]
$\pi_1 = 0.1986$, $\pi_2 = 0.2004 $, $\pi_3 = 0.1992$, $\pi_4 = 0.2015 $, $\pi_5 = 0.2002$
Notice that, when generate the data: each guassian components occupy 20% of all data. (1000 data point, 200 for each guassian components)

