Kaldi的在线自然梯度方法的算法实现
简介
神经网络的并行训练通常需要数据并行和模型并行。数据并行的常见方法涉及每个minibatch中模型参数的通信。该文献采用了不同的数据并行方法:在不同机器上运行多个SGD进程,每隔一段时间对模型参数进行平均并将平均后的模型参数分发到每台机器上。这对于大规模语音识别系统训练来说十分高效,但必须使用自然梯度随机梯度下降(natural gradient stochastic descent,NG-SGD)。
NaturalGradientAffineComponent
关键词:自然梯度下降、NG-SGD、自然梯度。
关于自然梯度的代码,位于kaldi/src/nnet3/nnet-simple-component.h以及nnet-precondition-online.h
NaturalGradientAffineComponent是AffineComponent的子类,重置的函数中最重要的是反向传播函数中参数更新的部分
src/nnet3/nnet-simple-component.cc |
void NaturalGradientAffineComponent::Update( const std::string &debug_info, const CuMatrixBase<BaseFloat> &in_value, const CuMatrixBase<BaseFloat> &out_deriv) {
CuMatrix<BaseFloat> in_value_temp; in_value_temp.Resize(in_value.NumRows(), in_value.NumCols() + 1, kUndefined); in_value_temp.Range(0, in_value.NumRows(), 0, in_value.NumCols()).CopyFromMat(in_value); in_value_temp.Range(0, in_value.NumRows(), in_value.NumCols(), 1).Set(1.0); CuMatrix<BaseFloat> out_deriv_temp(out_deriv);
BaseFloat in_scale, out_scale;
对输入矩阵X进行修正 preconditioner_in_.PreconditionDirections(&in_value_temp, &in_scale);
preconditioner_out_.PreconditionDirections(&out_deriv_temp, &out_scale); // "scale" is a scaling factor coming from the PreconditionDirections calls // (it's faster to have them output a scaling factor than to have them scale // their outputs). BaseFloat scale = in_scale * out_scale;
CuSubMatrix<BaseFloat> in_value_precon_part(in_value_temp, 0, in_value_temp.NumRows(), 0, in_value_temp.NumCols() - 1); // this "precon_ones" is what happens to the vector of 1's representing // offsets, after multiplication by the preconditioner. CuVector<BaseFloat> precon_ones(in_value_temp.NumRows()); precon_ones.CopyColFromMat(in_value_temp, in_value_temp.NumCols() - 1); BaseFloat local_lrate = scale * learning_rate_; bias_params_.AddMatVec(local_lrate, out_deriv_temp, kTrans, precon_ones, 1.0);
linear_params_.AddMatMat(local_lrate, out_deriv_temp, kTrans, in_value_precon_part, kNoTrans, 1.0); }
|
NaturalGradientRepeatedAffineComponent
src/nnet3/nnet-simple-component.cc |
void NaturalGradientRepeatedAffineComponent::Update( const CuMatrixBase<BaseFloat> &in_value, const CuMatrixBase<BaseFloat> &out_deriv) { KALDI_ASSERT(out_deriv.NumCols() == out_deriv.Stride() && in_value.NumCols() == in_value.Stride() && in_value.NumRows() == out_deriv.NumRows());
int32 num_repeats = num_repeats_, num_rows = in_value.NumRows(), block_dim_out = linear_params_.NumRows(), block_dim_in = linear_params_.NumCols();
CuSubMatrix<BaseFloat> in_value_reshaped(in_value.Data(), num_rows * num_repeats, block_dim_in, block_dim_in), out_deriv_reshaped(out_deriv.Data(), num_rows * num_repeats, block_dim_out, block_dim_out);
CuVector<BaseFloat> bias_deriv(block_dim_out); bias_deriv.AddRowSumMat(1.0, out_deriv_reshaped);
CuMatrix<BaseFloat> deriv(block_dim_out, block_dim_in + 1); deriv.ColRange(0, block_dim_in).AddMatMat( 1.0, out_deriv_reshaped, kTrans, in_value_reshaped, kNoTrans, 1.0); deriv.CopyColFromVec(bias_deriv, block_dim_in);
BaseFloat scale = 1.0; if (!is_gradient_) { try { // Only apply the preconditioning/natural-gradient if we're not computing // the exact gradient. 由于
} catch (...) { int32 num_bad_rows = 0; for (int32 i = 0; i < out_deriv.NumRows(); i++) { BaseFloat f = out_deriv.Row(i).Sum(); if (!(f - f == 0)) num_bad_rows++; } KALDI_ERR << "Preonditioning failed, in_value sum is " << in_value.Sum() << ", out_deriv sum is " << out_deriv.Sum() << ", out_deriv has " << num_bad_rows << " bad rows."; } } linear_params_.AddMat(learning_rate_ * scale, deriv.ColRange(0, block_dim_in)); bias_deriv.CopyColFromMat(deriv, block_dim_in); bias_params_.AddVec(learning_rate_ * scale, bias_deriv); } |
PerElementOffsetComponent
src/nnet3/nnet-simple-component.cc |
void PerElementOffsetComponent::Backprop( const std::string &debug_info, const ComponentPrecomputedIndexes *indexes, const CuMatrixBase<BaseFloat> &, // in_value const CuMatrixBase<BaseFloat> &, // out_value const CuMatrixBase<BaseFloat> &out_deriv, void *memo, Component *to_update_in, CuMatrixBase<BaseFloat> *in_deriv) const { PerElementOffsetComponent *to_update = dynamic_cast<PerElementOffsetComponent*>(to_update_in);
if (in_deriv && in_deriv->Data() != out_deriv.Data()) { // Propagate the derivative back to the input. in_deriv->CopyFromMat(out_deriv); }
if (to_update != NULL) { // we may have to reshape out_deriv, if "block-dim" was set // in the config file when initializing the object, leading // to dim_ being a multiple >1 of offset_.Dim(). // To avoid having separate code paths we create a sub-matrix // in any case, but this may just be a copy of out_deriv. int32 block_dim = offsets_.Dim(), multiple = dim_ / block_dim, block_stride = (multiple == 1 ? out_deriv.Stride() : block_dim), num_rows = out_deriv.NumRows() * multiple; KALDI_ASSERT(multiple == 1 || out_deriv.Stride() == out_deriv.NumCols()); CuSubMatrix<BaseFloat> out_deriv_reshaped(out_deriv.Data(), num_rows, block_dim, block_stride); if (!to_update->use_natural_gradient_ || to_update->is_gradient_) { KALDI_LOG << "Using non-NG update, lr = " << to_update->learning_rate_; to_update->offsets_.AddRowSumMat(to_update->learning_rate_, out_deriv_reshaped); } else { KALDI_LOG << "Using NG update, lr = " << to_update->learning_rate_; // make a copy as we don't want to modify the data of 'out_deriv', which // was const (even though CuSubMatrix does not respect const-ness in // this scenario) CuMatrix<BaseFloat> out_deriv_copy(out_deriv_reshaped); BaseFloat scale = 1.0; 由于
to_update->preconditioner_.PreconditionDirections(&out_deriv_copy, &scale); to_update->offsets_.AddRowSumMat(scale * to_update->learning_rate_, out_deriv_copy); } } } |
src/nnet3/nnet-convolutional-component.cc |
void TimeHeightConvolutionComponent::UpdateNaturalGradient( const PrecomputedIndexes &indexes, const CuMatrixBase<BaseFloat> &in_value, const CuMatrixBase<BaseFloat> &out_deriv) {
CuVector<BaseFloat> bias_deriv(bias_params_.Dim());
{ // this block computes 'bias_deriv', the derivative w.r.t. the bias. KALDI_ASSERT(out_deriv.Stride() == out_deriv.NumCols() && out_deriv.NumCols() == model_.height_out * model_.num_filters_out); CuSubMatrix<BaseFloat> out_deriv_reshaped( out_deriv.Data(), out_deriv.NumRows() * model_.height_out, model_.num_filters_out, model_.num_filters_out); bias_deriv.AddRowSumMat(1.0, out_deriv_reshaped); }
CuMatrix<BaseFloat> params_deriv(linear_params_.NumRows(), linear_params_.NumCols() + 1); params_deriv.CopyColFromVec(bias_deriv, linear_params_.NumCols());
CuSubMatrix<BaseFloat> linear_params_deriv( params_deriv, 0, linear_params_.NumRows(), 0, linear_params_.NumCols());
ConvolveBackwardParams(indexes.computation, in_value, out_deriv, 1.0, &linear_params_deriv);
梯度方向预调节 函数将输出一个标量,调节后的矩阵必须乘上该标量(这样可以省去一步CUDA运算)。 在对另一维运算进行自然梯度变换前,我们不会应用该缩放,否则该标量在多次迭代中会变得非常相似(尽管不同迭代间的标量不是不同的)
对二维矩阵形式的卷积核进行自然梯度变换时,将对2个维度先后进行变换。 BaseFloat scale1, scale2; 对第2个维度进行变换 preconditioner_in_.PreconditionDirections(¶ms_deriv, &scale1);
对第1个维度进行变换 CuMatrix<BaseFloat> params_deriv_transpose(params_deriv, kTrans); preconditioner_out_.PreconditionDirections(¶ms_deriv_transpose, &scale2);
linear_params_.AddMat( learning_rate_ * scale1 * scale2, params_deriv_transpose.RowRange(0, linear_params_.NumCols()), kTrans);
bias_params_.AddVec(learning_rate_ * scale1 * scale2, params_deriv_transpose.Row(linear_params_.NumCols())); } |
自然梯度方向预调节函数的具体实现解析
src/nnet3/natural-gradient-online.cc |
void OnlineNaturalGradient::PreconditionDirections( CuMatrixBase<BaseFloat> *X_t, BaseFloat *scale) { "梯度方向预调节"函数将矩阵的列作为目标维度进行 变换 if (X_t->NumCols() == 1) { // If the dimension of the space equals one then our natural gradient update // with rescaling becomes a no-op, but the code wouldn't naturally handle it // because rank would be zero. Support this as a special case. if (scale) *scale = 1.0; return; }
if (t_ == 0) // not initialized Init(*X_t);
int32 R = W_t_.NumRows(), D = W_t_.NumCols(); // space for W_t, J_t, K_t, L_t. CuMatrix<BaseFloat> WJKL_t(2 * R, D + R); WJKL_t.Range(0, R, 0, D).CopyFromMat(W_t_); BaseFloat rho_t(rho_t_); Vector<BaseFloat> d_t(d_t_);
bool updating = Updating();
BaseFloat initial_product;
initial_product = TraceMatMat(*X_t, *X_t, kTrans);
PreconditionDirectionsInternal(rho_t, initial_product, updating, d_t, &WJKL_t, X_t);
if (scale) { if (initial_product <= 0.0) { *scale = 1.0; } else { BaseFloat final_product = TraceMatMat(*X_t, *X_t, kTrans); *scale = sqrt(initial_product / final_product); } } t_ += 1; }
void OnlineNaturalGradient::PreconditionDirectionsInternal( const BaseFloat rho_t, const BaseFloat tr_X_Xt, bool updating, const Vector<BaseFloat> &d_t, CuMatrixBase<BaseFloat> *WJKL_t, CuMatrixBase<BaseFloat> *X_t) { int32 N = X_t->NumRows(), // Minibatch size. D = X_t->NumCols(), // Dimensions of vectors we're preconditioning R = rank_; // Rank of correction to unit matrix. KALDI_ASSERT(R > 0 && R < D); BaseFloat eta = Eta(N);
CuMatrix<BaseFloat> H_t(N, R); const CuSubMatrix<BaseFloat> W_t(*WJKL_t, 0, R, 0, D); // Below, WJ_t and LK_t are combinations of two matrices, // which we define in order to combine two separate multiplications into one. CuSubMatrix<BaseFloat> J_t(*WJKL_t, R, R, 0, D), L_t(*WJKL_t, 0, R, D, R), K_t(*WJKL_t, R, R, D, R), WJ_t(*WJKL_t, 0, 2 * R, 0, D), LK_t(*WJKL_t, 0, 2 * R, D, R);
H_t.AddMatMat(1.0, *X_t, kNoTrans, W_t, kTrans, 0.0); // H_t = X_t W_t^T
if (!updating) { // We're not updating the estimate of the Fisher matrix; we just apply the // preconditioning and return. // X_hat_t = X_t - H_t W_t
X_t->AddMatMat(-1.0, H_t, kNoTrans, W_t, kNoTrans, 1.0); return; }
J_t.AddMatMat(1.0, H_t, kTrans, *X_t, kNoTrans, 0.0); // J_t = H_t^T X_t
bool compute_lk_together = (N > D);
if (compute_lk_together) { // do the following two multiplies in one operation... // note // L_t = W_t J_t^T // K_t = J_t J_t^T // Note: L_t was defined as L_t = J_t W_t^T, but it's actually symmetric, // so we can compute it as L_t = W_t J_t^T.
LK_t.AddMatMat(1.0, WJ_t, kNoTrans, J_t, kTrans, 0.0); } else {
K_t.SymAddMat2(1.0, J_t, kNoTrans, 0.0); L_t.SymAddMat2(1.0, H_t, kTrans, 0.0); }
Matrix<BaseFloat> LK_cpu(LK_t); // contains L and K on the CPU. SubMatrix<BaseFloat> L_t_cpu(LK_cpu, 0, R, 0, R), K_t_cpu(LK_cpu, R, R, 0, R); if (!compute_lk_together) { // the SymAddMat2 operations only set the lower triangle and diagonal. L_t_cpu.CopyLowerToUpper(); K_t_cpu.CopyLowerToUpper(); }
// beta_t = \rho_t(1+\alpha) + \alpha/D tr(D_t) BaseFloat beta_t = rho_t * (1.0 + alpha_) + alpha_ * d_t.Sum() / D; Vector<BaseFloat> e_t(R), sqrt_e_t(R), inv_sqrt_e_t(R);
ComputeEt(d_t, beta_t, &e_t, &sqrt_e_t, &inv_sqrt_e_t); KALDI_VLOG(5) << "e_t = " << e_t;
// The double-precision Z_t here, and the scaling, is to avoid potential // overflow, because Z_t is proportional to the fourth power of data. SpMatrix<double> Z_t_double(R);
ComputeZt(N, rho_t, d_t, inv_sqrt_e_t, K_t_cpu, L_t_cpu, &Z_t_double); BaseFloat z_t_scale = std::max<double>(1.0, Z_t_double.Trace()); Z_t_double.Scale(1.0 / z_t_scale); SpMatrix<BaseFloat> Z_t_scaled(Z_t_double);
Matrix<BaseFloat> U_t(R, R); Vector<BaseFloat> c_t(R); 在CPU中进行对称特征值分解:
Kaldi::MatrixBase<BaseFloat>::Eig Z_t_scaled.Eig(&c_t, &U_t); SortSvd(&c_t, &U_t); c_t.Scale(z_t_scale);
const BaseFloat condition_threshold = 1.0e+06; // must_reorthogonalize will be true if the last diagonal element of c_t is // negative, since we don't take the absolute value, but this is the right // thing anyway. bool must_reorthogonalize = (c_t(0) > condition_threshold * c_t(R - 1));
BaseFloat c_t_floor = pow(rho_t * (1 - eta), 2); int32 nf; c_t.ApplyFloor(c_t_floor, &nf); if (nf > 0)
must_reorthogonalize = true; if (nf > 0 && self_debug_) { KALDI_WARN << "Floored " << nf << " elements of C_t."; }
X_t->AddMatMat(-1.0, H_t, kNoTrans, W_t, kNoTrans, 1.0); // X_hat_t = X_t - H_t W_t
Vector<BaseFloat> sqrt_c_t(c_t); sqrt_c_t.ApplyPow(0.5);
// \rho_{t+1} = 1/(D - R) (\eta/N tr(X_t X_t^T) + (1-\eta)(D \rho_t + tr(D_t)) - tr(C_t^{0.5})).
BaseFloat rho_t1 = 1.0 / (D - R) * (eta / N * tr_X_Xt + (1-eta)*(D * rho_t + d_t.Sum()) - sqrt_c_t.Sum()); // D_{t+1} = C_t^{0.5} - \rho_{t+1} I
Vector<BaseFloat> d_t1(sqrt_c_t);
d_t1.Add(-rho_t1);
BaseFloat floor_val = std::max(epsilon_, delta_ * sqrt_c_t.Max()); if (rho_t1 < floor_val)
rho_t1 = floor_val;
d_t1.ApplyFloor(floor_val);
CuMatrix<BaseFloat> W_t1(R, D); // W_{t+1}
ComputeWt1(N, d_t, d_t1, rho_t, rho_t1, U_t, sqrt_c_t, inv_sqrt_e_t, W_t, &J_t, &W_t1);
if (must_reorthogonalize) { if (self_debug_) { KALDI_WARN << "Reorthogonalizing."; } ReorthogonalizeRt1(d_t1, rho_t1, &W_t1, &J_t, &L_t); }
W_t_.Swap(&W_t1); d_t_.CopyFromVec(d_t1); rho_t_ = rho_t1;
if (self_debug_) SelfTest(); }
BaseFloat OnlineNaturalGradient::Eta(int32 N) const { if (num_minibatches_history_ > 0.0) { KALDI_ASSERT(num_minibatches_history_ > 1.0); return 1.0 / num_minibatches_history_; } else { KALDI_ASSERT(num_samples_history_ > 0.0); BaseFloat ans = 1.0 - exp(-N / num_samples_history_); // Don't let eta approach 1 too closely, as it can lead to NaN's appearing if // the input is all zero. if (ans > 0.9) ans = 0.9; return ans; } }
void OnlineNaturalGradient::ComputeZt(int32 N, BaseFloat rho_t, const VectorBase<BaseFloat> &d_t, const VectorBase<BaseFloat> &inv_sqrt_e_t, const MatrixBase<BaseFloat> &K_t, const MatrixBase<BaseFloat> &L_t, SpMatrix<double> *Z_t) const { // Use doubles because the range of quantities in Z_t can get large (fourth // power of data), and we want to avoid overflow. This routine is fast. BaseFloat eta = Eta(N); Vector<BaseFloat> d_t_rho_t(d_t);
d_t_rho_t.Add(rho_t); // now d_t_rho_t is diag(D_t + \rho_t I).
double etaN = eta / N, eta1 = 1.0 - eta, etaN_sq = etaN * etaN, eta1_sq = eta1 * eta1, etaN_eta1 = etaN * eta1; int32 R = d_t.Dim(); for (int32 i = 0; i < R; i++) {
double inv_sqrt_e_t_i = inv_sqrt_e_t(i), d_t_rho_t_i = d_t_rho_t(i); for (int32 j = 0; j <= i; j++) {
double inv_sqrt_e_t_j = inv_sqrt_e_t(j), d_t_rho_t_j = d_t_rho_t(j), L_t_i_j = 0.5 * (L_t(i, j) + L_t(j, i)), K_t_i_j = 0.5 * (K_t(i, j) + K_t(j, i)); // See (eqn:Zt) in header.
(*Z_t)(i, j) = etaN_sq * inv_sqrt_e_t_i * K_t_i_j * inv_sqrt_e_t_j + etaN_eta1 * inv_sqrt_e_t_i * L_t_i_j * inv_sqrt_e_t_j * d_t_rho_t_j + etaN_eta1 * d_t_rho_t_i * inv_sqrt_e_t_i * L_t_i_j * inv_sqrt_e_t_j + (i == j ? eta1_sq * d_t_rho_t_i * d_t_rho_t_i : 0.0); } } }
void OnlineNaturalGradient::ComputeWt1(int32 N, const VectorBase<BaseFloat> &d_t, const VectorBase<BaseFloat> &d_t1, BaseFloat rho_t, BaseFloat rho_t1, const MatrixBase<BaseFloat> &U_t, const VectorBase<BaseFloat> &sqrt_c_t, const VectorBase<BaseFloat> &inv_sqrt_e_t, const CuMatrixBase<BaseFloat> &W_t, CuMatrixBase<BaseFloat> *J_t, CuMatrixBase<BaseFloat> *W_t1) const {
int32 R = d_t.Dim(), D = W_t.NumCols(); BaseFloat eta = Eta(N); // \beta_{t+1} = \rho_{t+1} (1+\alpha) + \alpha/D tr(D_{t+1})
BaseFloat beta_t1 = rho_t1 * (1.0 + alpha_) + alpha_ * d_t1.Sum() / D; KALDI_ASSERT(beta_t1 > 0.0); Vector<BaseFloat> e_t1(R, kUndefined), sqrt_e_t1(R, kUndefined), inv_sqrt_e_t1(R, kUndefined); ComputeEt(d_t1, beta_t1, &e_t1, &sqrt_e_t1, &inv_sqrt_e_t1); Vector<BaseFloat> inv_sqrt_c_t(sqrt_c_t);
inv_sqrt_c_t.InvertElements();
Vector<BaseFloat> w_t_coeff(R); for (int32 i = 0; i < R; i++)
w_t_coeff(i) = (1.0 - eta) / (eta/N) * (d_t(i) + rho_t); CuVector<BaseFloat> w_t_coeff_gpu(w_t_coeff); // B_t = J_t + (1-\eta)/(\eta/N) (D_t + \rho_t I) W_t
J_t->AddDiagVecMat(1.0, w_t_coeff_gpu, W_t, kNoTrans, 1.0);
// A_t = (\eta/N) E_{t+1}^{0.5} C_t^{-0.5} U_t^T E_t^{-0.5}
Matrix<BaseFloat> A_t(U_t, kTrans); for (int32 i = 0; i < R; i++) {
BaseFloat i_factor = (eta / N) * sqrt_e_t1(i) * inv_sqrt_c_t(i); for (int32 j = 0; j < R; j++) {
BaseFloat j_factor = inv_sqrt_e_t(j);
A_t(i, j) *= i_factor * j_factor; } } // W_{t+1} = A_t B_t CuMatrix<BaseFloat> A_t_gpu(A_t);
W_t1->AddMatMat(1.0, A_t_gpu, kNoTrans, *J_t, kNoTrans, 0.0); }
void OnlineNaturalGradient::ComputeEt(const VectorBase<BaseFloat> &d_t, BaseFloat beta_t, VectorBase<BaseFloat> *e_t, VectorBase<BaseFloat> *sqrt_e_t, VectorBase<BaseFloat> *inv_sqrt_e_t) const { // e_{tii} = 1/(\beta_t/d_{tii} + 1) int32 D = d_t.Dim(); const BaseFloat *d = d_t.Data(); BaseFloat *e = e_t->Data(); for (int32 i = 0; i < D; i++)
e[i] = 1.0 / (beta_t / d[i] + 1); sqrt_e_t->CopyFromVec(*e_t);
sqrt_e_t->ApplyPow(0.5); inv_sqrt_e_t->CopyFromVec(*sqrt_e_t);
inv_sqrt_e_t->InvertElements(); }
void OnlineNaturalGradient::ReorthogonalizeRt1( const VectorBase<BaseFloat> &d_t1, BaseFloat rho_t1, CuMatrixBase<BaseFloat> *W_t1, CuMatrixBase<BaseFloat> *temp_W, CuMatrixBase<BaseFloat> *temp_O) { // threshold is a configuration value: a desired threshold on orthogonality, // below which we won't reorthogonalize. const BaseFloat threshold = 1.0e-03;
int32 R = W_t1->NumRows(), D = W_t1->NumCols();
BaseFloat beta_t1 = rho_t1 * (1.0 + alpha_) + alpha_ * d_t1.Sum() / D; Vector<BaseFloat> e_t1(R, kUndefined), sqrt_e_t1(R, kUndefined), inv_sqrt_e_t1(R, kUndefined);
ComputeEt(d_t1, beta_t1, &e_t1, &sqrt_e_t1, &inv_sqrt_e_t1);
temp_O->SymAddMat2(1.0, *W_t1, kNoTrans, 0.0); // O_{t+1} = E_{t+1}^{-0.5} W_{t+1} W_{t+1}^T E_{t+1}^{-0.5} Matrix<BaseFloat> O_mat(*temp_O); SpMatrix<BaseFloat> O(O_mat, kTakeLower); for (int32 i = 0; i < R; i++) {
BaseFloat i_factor = inv_sqrt_e_t1(i); for (int32 j = 0; j <= i; j++) {
BaseFloat j_factor = inv_sqrt_e_t1(j);
O(i, j) *= i_factor * j_factor; } }
if (O.IsUnit(threshold)) { if (self_debug_) { KALDI_WARN << "Not reorthogonalizing since already orthognoal: " << O; } return; } TpMatrix<BaseFloat> C(R); bool cholesky_ok = true; try { // one of the following two calls may throw an exception.
C.Cholesky(O);
C.Invert(); // Now it's C^{-1}.
if (!(C.Max() < 100.0)) { KALDI_WARN << "Cholesky out of expected range, " << "reorthogonalizing with Gram-Schmidt"; cholesky_ok = false; } } catch (...) { // We do a Gram-Schmidt orthogonalization, which is a bit less efficient but // more robust than the method using Cholesky. KALDI_WARN << "Cholesky or Invert() failed while re-orthogonalizing R_t. " << "Re-orthogonalizing on CPU."; cholesky_ok = false; } if (!cholesky_ok) {
Matrix<BaseFloat> cpu_W_t1(*W_t1); cpu_W_t1.OrthogonalizeRows();
W_t1->CopyFromMat(cpu_W_t1); // at this point cpu_W_t1 represents R_{t+1}- it has orthonormal // rows. Do: W_{t+1} = E_{t+1}^{0.5} R_{t+1}
CuVector<BaseFloat> sqrt_e_t1_gpu(sqrt_e_t1); W_t1->MulRowsVec(sqrt_e_t1_gpu); return; } // Next, compute (E_t^{0.5} C^{-1} E_t^{-0.5}) // but it's really t+1, not t.
for (int32 i = 0; i < R; i++) {
BaseFloat i_factor = sqrt_e_t1(i); for (int32 j = 0; j < i; j++) { // skip j == i because i_factor * j_factor == 1 for j == i.
BaseFloat j_factor = inv_sqrt_e_t1(j); C(i, j) *= i_factor * j_factor; } } O_mat.CopyFromTp(C); temp_O->CopyFromMat(O_mat); temp_W->CopyFromMat(*W_t1);
W_t1->AddMatMat(1.0, *temp_O, kNoTrans, *temp_W, kNoTrans, 0.0); }
|

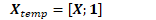





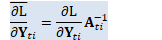



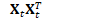
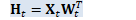
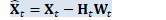
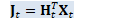
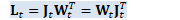
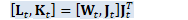
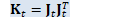
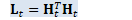
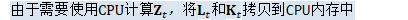
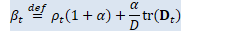
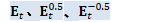
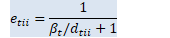
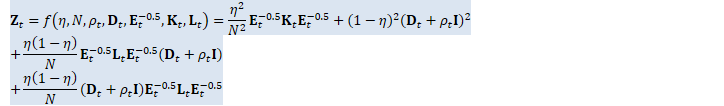
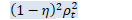

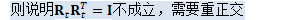
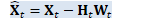



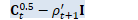



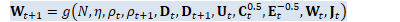
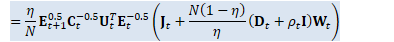
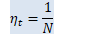

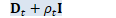

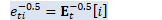
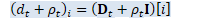
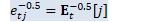

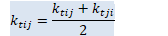
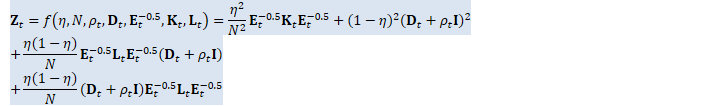
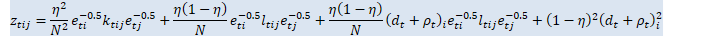
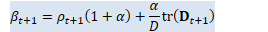
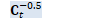
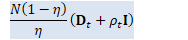
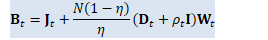

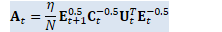

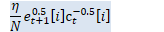
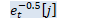

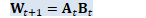
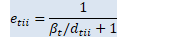

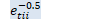
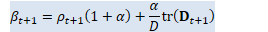
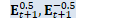





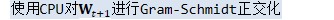

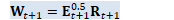
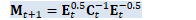
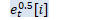

 浙公网安备 33010602011771号
浙公网安备 33010602011771号