【论文阅读】Simultaneously Self-Attending to All Mentions for Full-Abstract Biological Relation Extraction[NAACL]
原文链接:https://www.aclweb.org/anthology/N18-1080/
代码链接:https://github.com/patverga/bran
对关系抽取过程中的Mentions同时采用Self-Attending
论文目的:解决常年被忽视的跨句关系抽取问题,重新考虑mentions之间的交互interactions,提出了一个同时预测文档中所有提及对之间关系的模型。
Abstarct
We form pairwise predictions over entire paper abstracts using an efficient self-attention encoder.我们使用一个有效的自我注意编码器,对整篇论文抽象abstracts内容进行两两预测。Allpairs mention scores allow us to perform multi-instance learning by aggregating over mentions to form entity pair representations.所有成对的mentions分数允许我们通过聚合多个提及来进行多实例学习,从而形成实体对表示。We further adapt to settings without mention-level annotation by jointly training to predict named entities and adding a corpus of weakly labeled data.我们通过联合训练预测命名实体和添加弱标记数据的语料库,进一步适应没有提及级别注释的设置。在两个生物创造性基准数据集Biocreative benchmark datasets上的实验中,我们在没有外部知识库资源的模型上实现了生物创造性V化学疾病关系数据集Biocreative V Chemical Disease Relation dataset的最新性能。我们还引入了一个新的数据集,该数据集比现有的人类注释生物信息提取数据集大一个数量级,比远距监督的替代数据集更准确。
1 Introduction
In the CDR training set, this requires separately encoding and classifying each of the 5,318 candidate mention pairs independently, versus encoding each of the 500 abstracts once而不是对500个摘要每一个都编码一次.Though abstracts are longer than e.g. the text between mentions, many sentences contain multiple mentions, leading to redundant computation.
To facilitate efficient full-abstract relation extraction from biological text, we propose Bi-affine Relation Attention Networks (BRANs), a combination of network architecture, multi-instance and multi-task learning designed to extract relations between entities in biological text without requiring explicit mention-level annotation.为了从生物文本中有效提取全抽象的关系,我们提出了双仿射关系注意网络(BRANs),这是一种结合网络结构、多实例和多任务学习的方法,旨在提取生物文本中实体之间的关系,而不需要明确的提及级注释。【远程监督】We synthesize convolutions and self-attention, a modification of the Transformer encoder introduced by Vaswani et al. (2017), over sub-word tokens to efficiently incorporate into token representations rich context between distant mention pairs across the entire abstract.作者从transformer综合了convoltuions和self-attention,通过子词sub-word tokens,有效地将整个抽象abstract中遥远提及对之间的丰富上下文context合并incorporate到tokens表示中。We score all pairs of mentions in parallel using a bi-affine operator, and aggregate over mention pairs using a soft approximation of the max function in order to perform multi-instance learning.使用bi-affine 为所有mention对进行评分,并使用max函数的软近似值聚合所有提及对以为了进行多实例学习。
在两个基准的生物关系提取数据集上的大量实验中,我们在Biocreative V CDR数据集上的实验中,在不使用外部知识库资源的情况下,实现了最先进的模型性能,并在Biocreative VI ChemProt数据集上优于可比基线。我们还引入了一个新的数据集,该数据集比现有的黄金标记生物关系提取数据集大一个数量级,同时覆盖了更广泛的实体和关系类型,比相同大小的远距监督数据集具有更高的精度。我们在这个新的数据集上提供了一个强大的基线,并鼓励将其作为未来生物关系提取系统的基准。
2 Model
我们设计我们的模型,以有效地编码跨多个句子的长上下文,同时形成成对的预测,而不需要提到成对特定的特征without the need for mention pair-specific features。为此,我们的模型首先使用自我注意self-attention对输入令牌嵌入input tokens embeddigs进行编码。这些嵌入用于预测实体和关系。关系提取模块将每个tokens转换为头head和tail表示。These representations are used to form mention pair predictions using a bi-affine operation with respect to learned relation embeddings.这些表示被用于使用bi-affine 操作得到一个有训练过的关系嵌入的提及对预测。最后,这些提及对预测被汇集起来形成实体对预测,表示每个关系类型是否由每个关系对表示expressing whether each relation type is expressed by each relation pair。
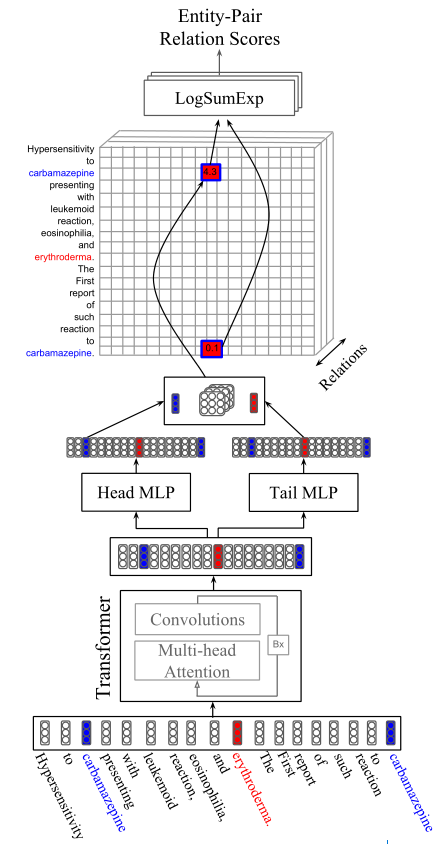
Figure 1: The relation extraction architecture. Inputs are contextually encoded using the Transformer(Vaswani et al., 2017), made up of $B$ layers of multi-head attention and convolution subcomponents. Each transformed token is then passed through a $head$ and $tail$ MLP to produce two position-specific representations. A bi-affine operation is performed between each $head$ and $tail$ representation with respect to each relation’s embedding matrix, producing a pair-wise relation affinity tensor. Finally, the scores for cells corresponding to the same entity pair are pooled with a separate LogSumExp operation for each relation to get a final score. The colored tokens illustrate calculating the score for a given pair of entities; the model is only given entity information when pooling over mentions.关系提取体系结构。输入使用Transformer进行上下文编码,由B层多头注意和卷积子组件组成。然后,将每个转换后的tokens通过head and tail MLP传递,以产生两种特定于位置的表示。针对每个关系的嵌入矩阵,在每个头和尾表示之间执行一个双仿射操作,产生一对关系亲和张量pair-wise relation affinity tensor。最后,将对应于同一实体对的单元格cells的分数与每个关系的单独LogSumExp操作进行池化,以获得最终分数。彩色标记说明了计算给定一对实体的分数;模型只在mentions上的池化操作时给出实体信息。
2.1 Inputs
Our model takes in a sequence of $N$ token embeddings in $R^d$.Because the Transformer has no innate notion of token position, the model relies on positional embeddings which are added to the input token embeddings
因为Transformer没有token位置的固有概念,所以模型依赖于添加到输入令牌嵌入input token embeddings中的位置嵌入。We learn the position embedding matrix $P^{m×d}$ which contains a separate $d$ dimensional embedding for each position, limited to $m$ possible positions.Our final input representation for token $x_i$ is:
$x_i=s_i+p_i$
where $s_i$ is the token embedding for $x_i$ and $p_i$ is the positional embedding for the $i$th position. If $i$ exceeds $m$, we use a randomly initialized vector in place of $p_i$.
We tokenize the text using byte pair encoding (BPE) (Gage, 1994; Sennrich et al., 2015). The BPE algorithm constructs a vocabulary of sub-word pieces, beginning with single characters. Then, the algorithm iteratively merges the most frequent cooccurring tokens into a new token, which is added to the vocabulary. This procedure continues until a pre-defined vocabulary size is met.我们使用字节对编码(BPE)对文本进行标记化。BPE算法从单个字符开始构建子单词片段的词汇表。然后,该算法迭代地将最频繁出现的tokens合并成一个新的tokens,并将其添加到词汇表中。此过程将继续,直到满足预定义的词汇表大小。
BPE is well suited for biological data for the following reasons. First, biological entities often have unique mentions made up of meaningful subcomponents, such as 1,2-dimethylhydrazine. Additionally, tokenization of chemical entities is challenging, lacking a universally agreed upon algorithm (Krallinger et al., 2015). As we demonstrate in §3.3.2, the subword representations produced by BPE allow the model to formulate better predictions, likely due to better modeling of rare and unknown words.BPE非常适合于生物数据,原因如下。首先,生物实体通常由有意义的亚组分组成,例如1,2-二甲基肼。此外,化学实体的标记化具有挑战性,缺乏普遍认可的算法(Krallinger et al.,2015)。正如我们在§3.3.2中所展示的,BPE产生的子词表示允许模型制定更好的预测,这可能是由于对罕见和未知词的更好建模。
2.2 Transformer
We base our token encoder on the Transformer self-attention model (Vaswani et al., 2017). The Transformer is made up of B blocks. Each Transformer block, which we denote $Transformer_k$, has its own set of parameters and is made up of two subcomponents子组件: multi-head attention and a series of convolutions【The original Transformer uses feed-forward connections, i.e. width-1 convolutions, whereas we use convolutions with width > 1卷积宽度改变】The output for token $i$ of block $k$, $b_i^{(k)}$, is connected to its input $b_i^{(k-1)}$ with a residual connection 残差链接(He et al., 2016). Starting with $b_i^{(0)}=x_i$:
$b_i^{(k)}=b_i^{(k-1)}+Transformer_k(b_i^{(k-1)})$
2.2.1 Multi-head Attention
Multi-head attention applies self-attention multiple times over the same inputs using separately normalized parameters (attention heads) and combines the results, as an alternative to applying one pass of attention with more parameters.多头注意多个头都分别使用标准化参数在相同的输入上多次self-attention,并将结果结合起来,作为应用一次注意和更多参数的替代方法。这种建模决策背后的直觉是,将注意力划分为多个头部,使模型更容易学会用每个头部关注不同类型的相关信息。The self-attention updates input $b_i^{(k−1)}$ by performing a weighted sum over all tokens in the sequence, weighted by their importance for modeling token $i$,自我注意通过对序列中的所有令牌执行加权和来更新输入$b_i^{(k-1)}$,加权和由它们对令牌$i$建模的重要性来加权。
Each input is projected to a key $k$, value $v$, and query $q$, using separate affine transformations with ReLU activations (Glorot et al., 2011).每个输入都被投影到一个键$k$、值$v$和查询$q$,使用单独的仿射变换和ReLU激活。Here, $k$, $v$, and $q$ are each in$R^{\frac{d}{H}}$ where $H$ is the number of heads. The attention weights $a_{ijh}$for head $h$ between tokens $i$ and $j$ are computed using scaled dot-product attention:
$a_{ijh}=\sigma(\frac{q_{ih}^Tk_{jh}}{\sqrt{d}})$
$o_{ih}=\sum_jv_{jh}\bigodot a_{ijh}$
$\bigodot$denoting element-wise multiplication,$\sigma$ indicating a softmax along the $j$th dimension,σ表示沿第$j$维的软最大值,The scaled attention is meant to aid optimization by flattening the softmax and better distributing the gradients (Vaswani et al., 2017).按比例关注旨在通过使softmax平坦化和更好地分布梯度来帮助优化。【使用softmax的好处】
The outputs of the individual attention heads are concatenated, denoted $[·;·]$, into $o_i$. All layers in the network use residual connections between the output of the multi-headed attention and its input. Layer normalization (Ba et al., 2016), denoted $LN(·)$, is then applied to the output.
$o_i=[o_1;...;o_h]$
$m_i=LN(b_i^{(k-1)}+o_i)$
2.2.2 Convolutions
The second part of our Transformer block is a stack of convolutional layers. The sub-network used in Vaswani et al. (2017) uses two width-1 convolutions.We add a third middle layer with kernel width 5, which we found to perform better. Many relations are expressed concisely简洁地 by the immediate local context,e.g. Michele’s husband Barack, or labetalol-induced hypotension. Adding this explicit n-gram modeling is meant to ease the burden on the model to learn to attend to local features.添加这种显式的n-gram建模是为了减轻模型学习处理局部特征的负担。We use $C_w(·)$ to denote a convolutional operator with kernel width $w$. Then the convolutional portion of the transformer block is given by:
$t_i^{(0)}=ReLU(C_1(m_i))$
$t_i^{(1)}=ReLU(C_5(t_i^{(0)}))$
$t_i^{(2)}=C_1(t_i^{(1)})$
Where the dimensions of $t_i^{(0)}$ and $t_i^{(1)}$ are in $R^{4d}$ and that of $t_i^{(2)}$ is in $R^d$.
2.3 Bi-affine Pairwise Scores
We project each contextually encoded token $b_i^{(B)}$ through two separate MLPs to generate two new versions of each token corresponding to whether it will serve as the first (head) or second (tail) argument of a relation:
$e_i^{head}=W_{head}^{(1)}(ReLU(W_{head}^{(0)}b_i^{(B)}))$
$e_i^{tail}=W_{tail}^{(1)}(ReLU(W_{tail}^{(0)}b_i^{(B)}))$
We use a bi-affine operator to calculate an $N×L×N$ tensor $A$ of pairwise affinity scores, scoring each (head, relation, tail) triple:
$A_{ilj}=(e_i^{head}L)e_j^{tail}$
where L is a $d×L×d$ tensor, a learned embedding matrix for each of the $L$ relations.In subsequent sections we will assume we have transposed the dimensions of A as $d × d × L$ for ease of indexing.
2.4 Entity Level Prediction
Our data is weakly labeled in that there are labels at the entity level but not the mention level, making the problem a form of strong-distant supervision (Mintz et al., 2009)【有实体级标签,无提及级标签】。In distant supervision, edges in a knowledge graph are heuristically applied to sentences in an auxiliary unstructured text corpus — often applying the edge label to all sentences containing the subject and object of the relation在远程监控中,知识图中的边启发式地应用于辅助非结构化文本语料库中的句子——通常将边标签应用于包含关系主语和宾语的所有句子。由于该过程不精确,并且在训练数据中引入了噪声,因此引入了多实例学习等方法(Riedel et al.,2010;Surdeanu et al.,2012)。在多实例学习中,模型不是孤立地观察每个远距离标记的提及对,而是对这些提及的集合进行训练,并进行单个更新。More recently, the weighting function of the instances has been expressed as neural network attention (Verga and McCallum, 2016; Lin et al., 2016; Yaghoobzadeh et al., 2017).【network attention频繁出现】。
We aggregate over all representations for each mention pair in order to produce per-relation scores for each entity pair.我们对每个提及对的所有表示进行聚合,以便为每个实体对生成每个关系分数。For each entity pair $(p^{head}, p^{tail})$, let $P^{head}$ denote the set of indices of mentions of the entity $p^{head}$, and let $P^{tail}$ denote the indices of mentions of the entity $p^{tail}$.Then we use the LogSumExp function to aggregate the relation scores from $A$ across all pairs of mentions of $p^{head}$ and $p^{tail}$:
$scores(p^{head},p^{tail})=log\sum_{i∈P^{head},j∈P^{tail}}exp(A_{ij})$
The LogSumExp scoring function is a smooth approximation to the max function and has the benefits of aggregating information from multiple predictions and propagating dense gradients as opposed to the sparse gradient updates of the max (Das et al., 2017).LogSumExp评分函数是max函数的平滑近似,具有从多个预测聚合信息和传播密集梯度(与max的稀疏梯度更新相反)的优点。
2.5 Named Entity Recognition
In addition to pairwise relation predictions, we use the Transformer output $b_i^{(B)}$ to make entity type predictions. We feed $b_i^{(B)}$ as input to a linear classifier which predicts the entity label for each token with per-class scores $c_i$除了成对的关系预测外,我们还使用Transformer输出$b_i^{(B)}$来进行实体类型预测。我们将$b_i^{(B)}$作为输入输入到一个线性分类器,它用每个类的分数$c_i$预测每个标记的实体标签:
$c_i=W^{(3)}b_i^{(B)}$
We augment the entity type labels with the BIO encoding to denote entity spans. We apply tags to the byte-pair tokenization by treating each subword within a mention span as an additional token with a corresponding B- or I- label.我们使用BIO编码来增加实体类型标签,以表示实体跨度。我们将标记应用于字节对标记化,方法是将提及范围内的每个子词视为具有相应B-或I-标签的额外标记。
2.6 Training
We train both the NER and relation extraction components of our network to perform multi-class classification using maximum likelihood, where NER classes $y_i$ or relation classes $r_i$ are conditionally independent given deep features produced by our model with probabilities given by the softmax function. In the case of NER, features are given by the per-token output of the transformer我们训练网络的NER和关系提取组件,使用最大似然来执行多类分类,其中NER类和关系类是条件独立的,因为我们的模型产生了深度特征,概率由softmax函数给出。对于NER,features由Transformer的每个令牌输出给出:
$\frac{1}{N}\sum_{i=1}^NlogP(y_i|b_i^{(B)})$
In the case of relation extraction, the features for each entity pair are given by the LogSumExp over pairwise scores described in § 2.4. For E entity pairs, the relation $r_i$ is given by
在关系抽取的情况下,每个实体对的特性由LogSumExp在§2.4中描述的成对得分给出。对于$E$个实体对,关系$r_i$为:
$\frac{1}{E}\sum_{i=1}^ElogP(r_i|scores(p^{head},p^{tail}))$
We train the NER and relation objectives jointly, sharing all embeddings and Transformer parameters. To trade off the two objectives, we penalize the named entity updates with a hyperparameter $\lambda$我们共同训练NER和关系目标,共享所有的嵌入和变压器参数。为了平衡这两个目标,我们用超参数惩罚命名实体更新。
3 Results
我们在三个数据集上评估我们的模型:生物创新V化学疾病关系基准Biocreative V Chemical Disease Relation benchmark(CDR),它为化学物质和疾病之间的关系建模(§3.1);Biocreative VI ChemProt基准Biocreative VI ChemProt benchmark(CPR),它模拟了化学品和蛋白质之间的关系(§3.2);以及我们在§3.3中描述的基于化学毒理学数据库Biocreative VI ChemProt benchmark(CTD)的人类管理的新的、大的和准确的数据集,该数据库模拟了化学物质、蛋白质和基因之间的关系。
CDR数据集在论文摘要abstracts的层次上进行注释,需要考虑到远距离、跨句关系,因此对该数据集的评估表明我们的模型能够进行这样的推理。我们还通过对CPR数据集进行实验来评估我们的模型在更传统的设置(不需要跨句子建模)中的性能,因为所有注释都位于单个句子中提到的两个实体之间。最后,我们提出了一个使用强远程监督(§2.4)构建的新数据集,并在文档级别上添加注释。这个数据集比其他数据集大得多,包含更多的关系类型,并且需要在句子之间进行推理。
CTD注释仅在文档级别,不包含提及注释
The CDR dataset is a subset of these original annotations, supplemented with human annotated, entity linked mention annotationsCDR数据集是这些原始注释的子集,并添加了人工注释、实体链接的提及注释,此数据集中的关系注释也仅在文档级别。

在表2中,我们展示了在没有使用语言特征的情况下优于基线的结果。我们展示了20个随机种子的平均性能超过20次,以及他们平均预测的集合。通过添加弱标记数据,我们看到了性能的进一步提升。
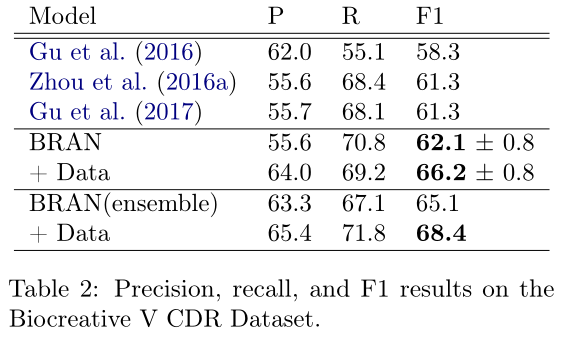
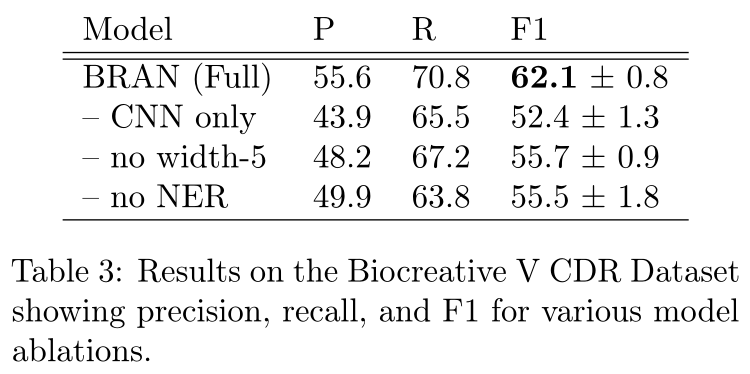
表3显示了我们模型中消融部分的效果。“CNN only”从变压器块中移除了多头注意力组件,“no width-5”用width-1卷积替换了变压器前馈组件的width-5卷积,“no NER”删除了命名实体识别多任务目标(§2.5)。
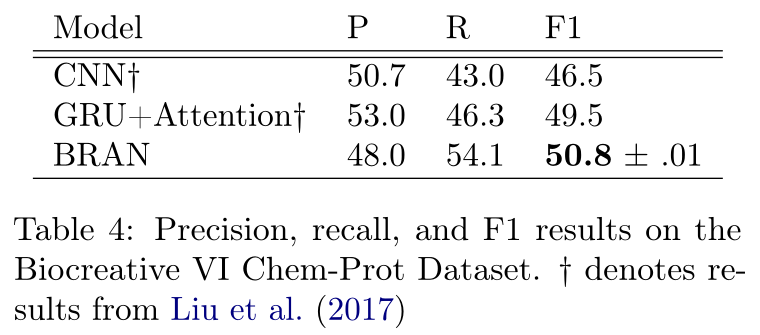
随后作者为了评估模型在没有明确评估跨句关系的环境中的表现,在Biocreative VI ChemProt数据集(CDR)上进行了实验
在表4中,我们看到尽管我们的模型同时形成了句子中所有实体对之间的所有预测,但我们能够超越独立分类每个提及对的先进模型。有趣的是,我们的模型具有较高的查全率和较低的查准率,而基线模型都具有精度偏差,查全率较低。这表明,结合这些风格的模型可以在这项任务上取得进一步的进展。
作者觉得现有远程监督数据集存在一些问题。远程监控数据集对所有语句应用独立于文档的、完整级别的注释,从而导致很大一部分不正确的标签。对这些数据的评估涉及非常小的(几百个)带黄金注释的例子或交叉验证,以预测噪音较大、应用较远的标签。优化很多过程自制了一个数据集,过程我就略了。
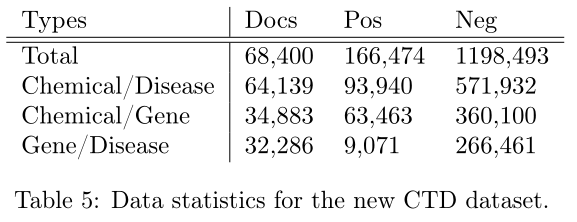
细化过后的新数据集关系类型:
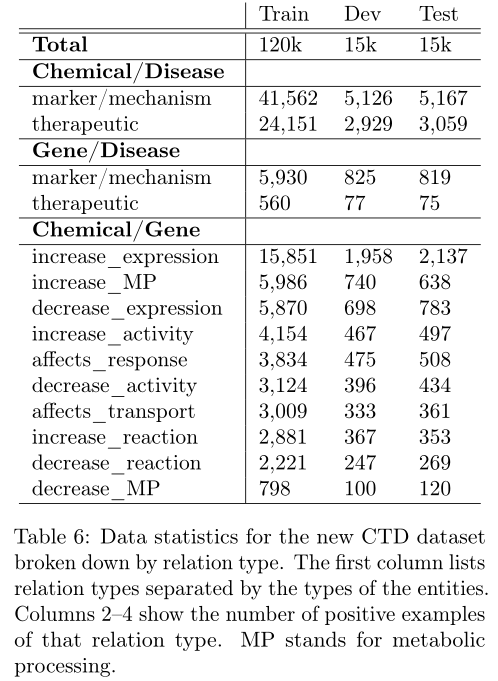
Figure 2 depicts the model’s performance on relation extraction as a function of distance between entities. For example, the blue bar depicts performance when removing all entity pair candidates (positive and negative) whose closest mentions are more than 11 tokens apart. We consider removing entity pair candidates with distances of 11, 25, 50, 100 and 500 (the maximum document length). The average sentence length is 22 tokens. We see that the model is not simply relying on short range relationships, but is leveraging information about distant entity pairs, with accuracy increasing as the maximum distance considered increases. Note that all results are taken from the same model trained on the full unfiltered training set.图2描述了作为实体之间距离函数的模型在关系提取方面的性能。例如,蓝色条描述了移除所有实体对候选者(正和负)时的性能,这些候选者最近的提及间隔超过11个标记。我们考虑删除距离为11、25、50、100和500(最大文档长度)的实体对候选。平均句子长度为22个标记。我们看到,该模型不仅仅依赖于短程关系,而是利用了有关远程实体对的信息,随着所考虑的最大距离的增加,精确度也随之增加。请注意,所有结果都来自在完整的未过滤训练集上训练的同一模型。
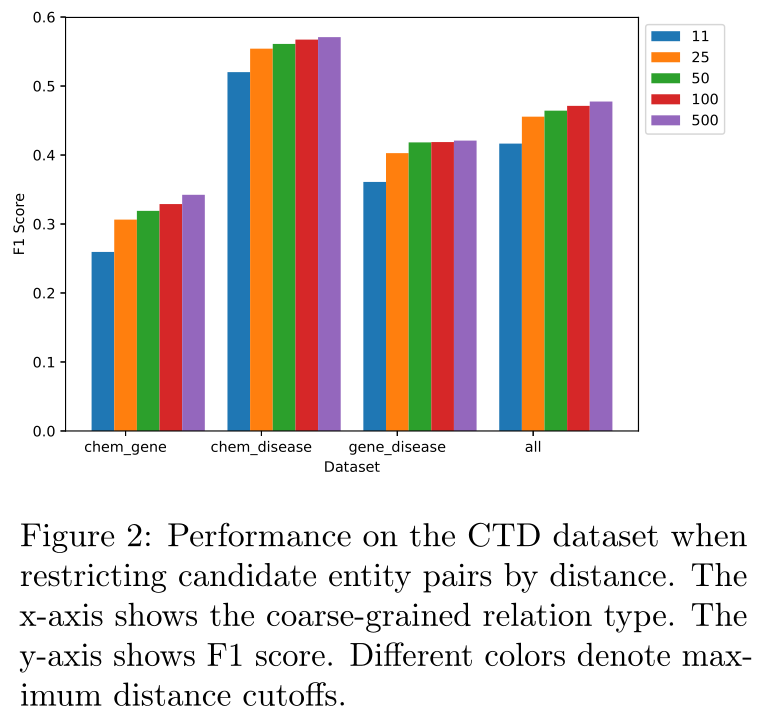
图2:按距离限制候选实体对时CTD数据集的性能。x轴显示粗粒度关系类型。y轴显示F1分数。不同的颜色表示最大距离截止。
模型在full CTD:
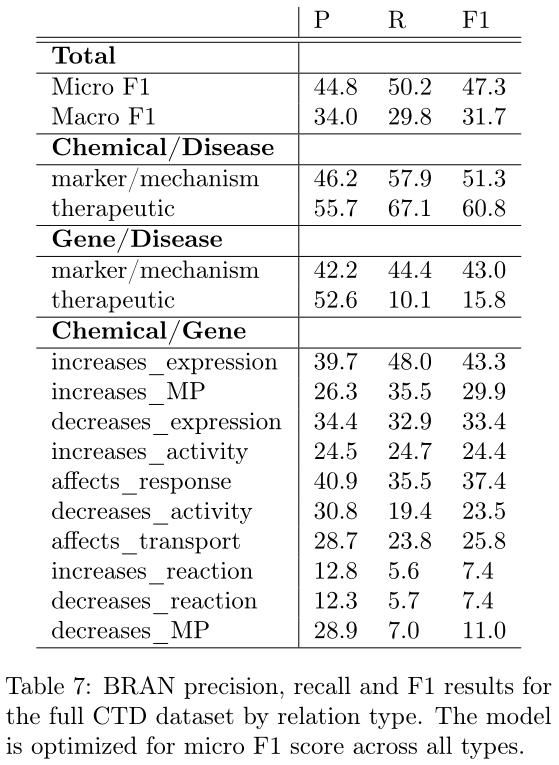
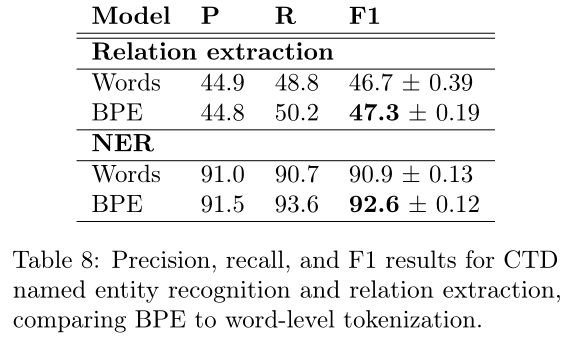
使用BPE方法经验证非常适合生物数据:
5 Conclusion
我们提出一个双仿射关系注意网络bi-affine relation attention network,同时为文档中的所有提及对打分。我们的模型在三个数据集上表现良好,包括两个标准的基准生物关系提取数据集和一个新的、大的和高质量的数据集。尽管我们没有额外的语言资源,也没有提到特定的配对pair-specific特征,但是我们的模型在生物创造性的V-CDR数据集上的表现超过了以前的技术水平。
我们目前的模型只能预测数据给出的固定关系模式。然而,这可以通过将我们的模型集成到开放关系提取架构中来改善by integrating our model into open relation extraction architectures such as Universal Schema (Riedel et al., 2013; Verga et al., 2016b))。我们的模型也适用于其他成对评分任务,如超词预测、共同引用解析和实体解析。我们将在今后的工作中研究这些方向。



 浙公网安备 33010602011771号
浙公网安备 33010602011771号